Surface proteins of Propionibacterium freudenreichii are involved in its anti-inflammatory properties
Propionibacterium freudenreichii is a beneficial bacterium used in the food industry as a vitamin producer, as a bio-preservative, as a cheese ripening starter and as a probiotic.
Propionibacterium freudenreichii is a beneficial bacterium used in the food industry as a vitamin producer, as a bio-preservative, as a cheese ripening starter and as a probiotic. It is known to adhere to intestinal epithelial cells and mucus and to modulate important functions of the gut mucosa, including cell proliferation and immune response. Adhesion of probiotics and cross-talk with the host rely on the presence of key surface proteins, still poorly identified. Identification of the determinants of adhesion and of immunomodulation by P. freudenreichii remains a bottleneck in the elucidation of its probiotic properties. In this report, three complementary proteomic methods are used to identify surface-exposed proteins in a strain, previously selected for its probiotic properties. The role of these proteins in the reported immunomodulatory properties of P. freudenreichii is evidenced. This work constitutes a basis for further studies aimed at the elucidation of mechanisms responsible for its probiotic effects, in a post-genomic context.
Biological significance
Dairy propionibacteria, mainly the species Propionibacterium freudenreichii, are consumed in high amounts within Swiss type cheeses. These peculiar bacteria are considered both as dairy starters and as probiotics. Their consumption modulates the gut microbiota, which makes them both probiotic and prebiotic. Promising immunomodulatory properties have been identified in these bacteria, in vitro, in animals and in humans. However, the mechanisms responsible for such anti-inflammatory properties are still unknown. In this work, we identify surface proteins involved in adhesion and immunostimulation by P. freudenreichii. This opens new perspectives for its utilization in new functional fermented food products, in clinical trials, and in understanding modulation of gut inflammation by products containing propionibacteria.
© 2014 Elsevier B.V. All rights reserved
1. Introduction
Propionibacterium freudenreichii is a GRAS (Generally Recognized As Safe) actinobacterium consumed in high amounts in our diet. It is traditionally used in the food industry as a vitamin producer, as a bio-preservative and as a cheese ripening starter.
Although less studied than lactobacilli or bifidobacteria, dairy propionibacteria, mainly P. freudenreichii, recently attracted attention because of their unique probiotic potential. They are included in commercial probiotic preparations, available as tablets or capsules, intended to improve intestinal transit and comfort. Probiotics are defined as “living microorganisms which when administered in adequate amounts confer a health benefit on the host” [1]. P. freudenreichii consumption modulates the human complex intestinal microbiota by enhancing bifidobacteria population [2–8]. Furthermore, in vitro and in vivo experimental data suggest a protective role of P. freudenreichii metabolites, including short chain fatty acids, in the context of carcinogenesis, by favoring apoptotic depletion of colon cancer cells [9–11]. Selected strains of P. freudenreichii were also shown to exert immunomodulation with anti-inflammatory effects confirmed in vivo [8,12,13]. By contrast, a subset of P. freudenreichii strains, covered with an extracellular polysaccharide capsule, displays no immunomodulatory property. Suppression of this capsule by gene inactivation confers immunomodulatory properties to these strains [14]. Probiotic effect within the gut is favored by the great hardiness of P. freudenreichii, which adapts to various harsh conditions [15], including the human gut [16], in accordance with the remarkable stress-adaptability suggested by the genome of the CIRM-BIA1 type strain [17]. Its annotation revealed redundant stress adaptation machinery including molecular chaperones, proteases, thioredoxin systems, bile acid and multidrug resistance transporters. Local effect of propionibacterial beneficial metabolites, as well as immunomodulation, is also favored by the ability of P. freudenreichii to adhere to host cells and mucus [18–21], a property which depends on surface proteins that are still not identified.
Adhesion to host cells and mucus, survival within the gut and interaction (cross-talk) with the host are key properties of probiotic bacteria, which may be correlated [22,23]. They depend on key surface compounds, including surface proteins [24]. Deciphering the surface proteome of probiotic bacteria thus constitutes a hot research area which will participate in the elucidation of key mechanisms underlying the bacterium/ host cross-talk [25]. Bacterial genome sequences being increasingly available, in silico prediction of proteins' surface exposure is now made possible by dedicated software for Gram-positive bacteria, in particular. Proteins are predicted as cytoplasmic, membrane, cell wall, or secreted, based on in silico detection of signal peptides, either from secreted proteins or from membrane bound lipoproteins [26], hydrophobic trans-membrane segments, or conserved domains or motifs indicative of proteins covalently or non-covalently linked to the cell wall [27]. The recently developed SurfG+ sequence analysis software also takes into consideration integral membrane proteins exposing specific parts at the surface of the bacterium [28]. Such in silico approach should be used in conjunction with “wet lab” proteomic tools.
The first proteomic investigations of probiotic bacteria dealt with cytoplasmic or whole-cell protein extracts and were limited by the incompatibility of some cell wall and/or surface proteins with two-dimensional electrophoresis, due to their size, isoelectric point and poor solubility. The second bottleneck was the lack of available sequenced and annotated genomes allowing the identification of proteins using proteomic tools such as sequencing or mass spectrometry. Another limit was the lack of a method for specifically detecting surface proteins. Gel-based approaches, as well as novel gel-free ones, are currently focusing on cell surface proteins.
Selective extraction of surface proteins from intact bacteria using chaotropic agents such as LiCl has been used to identify cell wall surface proteins of Lactobacillus acidophilus [29], including the surface layer protein SlpA, which is involved in the cytokine response elicited by L. acidophilus [30]. Such procedures are restricted to proteins non-covalently anchored to cell wall polymers via electrostatic interactions, involving SLH (S-Layer Homology) domains [31,32]. More recently, a gel-free enzymatic method consisting of the enzymatic “shaving” of surface proteins with a proteolytic enzyme, most often trypsin, followed by the identification of released peptides using liquid chromatography coupled to mass spectrometry (LC–MS/MS), was developed. It has been successfully used to identify surface proteins in the pathogens Streptococcus pyogenes [33,34], Staphylococcus pseudintermedius and Staphylococcus aureus [35], in the commensal/opportunistic pathogen Enterococcus faecalis [36,37] and in the dairy starter Lactococcus lactis [38]. It should be noticed that surface proteins that do not expose an accessible trypsin cleavage site at the surface are not identified this way, and that released peptides with covalent modifications, such as complex glycosylation, may not be identified by LC–MS/MS. Finally, selective labeling of surface accessible proteins using in situ covalent binding of a fluorescent dye, CyDye (usually used in 2D DIGE experiments), was also described. It was first developed for cultured eukaryotic cells [39] and then adapted to the surface proteome of the pathogenic mollicute Mycoplasma genitalium [40]. This method does not depend on surface accessible trypsin cleavage sites but on the common amine functions of aminoacid side chains, reacting with the CyDye NHS ester reactive group. It however depends on the limits of the separation of proteins by two-dimensional electrophoresis, including the pH gradient used. Different methods having different drawbacks, combining the 3 approaches should maximize the accuracy of surface protein identification.
No experimentally-supported inventory of P. freudenreichii surface proteins is, to our knowledge, available to date. However, its surface components are likely to play a role in its interaction with the environment [14]. This includes the dairy matrix in which it grows in fermented dairy products. Such interaction is strongly suggested by the observed preferential localization of propionibacteria at the fat/protein interface in Emmental cheese [41]. The reported adhesion to intestinal epithelial cells and mucus also suggests the involvement of surface adhesins [42,43].
Moreover, extraction of surface proteins using guanidine hydrochloride abolished the immunomodulatory properties of several P. freudenreichii strains [13]. Neither enzymatic shaving, nor CyDye labeling has yet been applied to beneficial probiotic bacteria, including P. freudenreichii.
In this work, we have selected an ITG P20 strain of P. freudenreichii, also called CIRM-BIA 129, which is used as a cheese ripening starter [44,45]. It also displays promising probiotic traits, particularly immunomodulation, and was spotted as the most effective strain in inducing the IL-10 regulatory cytokine [12]. The genome of this strain was sequenced and annotated using the Agmial platform [46]. The subcellular localization of the proteins encoded by this genome was predicted using SurfG+ [28]. In this work, we have combined extraction, shaving and labeling to inventory its surface proteins. This work constitutes the first experimental inventory of P. freudenreichii surface proteins and reveals proteins known, in other microorganisms, to participate in bacterium/host interactions.
2. Materials and methods
2.1. Bacterial cultures in a dairy medium
P. freudenreichii strain ITG P20, also called CIRM-BIA 129, isolated by Actalia Dairy Products (Institut Technique du Gruyère, Actalia, Rennes, France), was provided by the CIRM-BIA Biological Resource Center (Centre International de Ressources Microbiennes-Bactéries d'Intérêt Alimentaire, INRA, Rennes, France). It was cultivated at 30 °C without shaking in cow's milk ultrafiltrate supplemented with 50 mM of sodium L-lactate(galaflow SL60, Société Arnaud, Paris, France) and 5 g/L of casein hydrolysate (Organotechnie, La Courneuve, France), sterilized by 0.2 μm filtration (Nalgene, Roskilde, Denmark) as described previously [47]. Milk ultrafiltrate was produced using a UF pilot equipment (T.I.A., Bollene, France) equipped with an organic spiral membrane with a molecular weight cut-off of 5 kDa (Koch International, Lyon, France). Growth was monitored spectrophotometrically at 650 nm (OD650), as well as by counting
colony-forming units (CFUs) in YEL medium [48] containing 1.5% agar. Bacteria were harvested in a stationary phase (76 h, 109 CFU/mL) by centrifugation (6000 ×g, 10 min, 4 °C).
2.2. Bio-informatics
The genome of the ITG P20 alias CIRM BIA 129 strain was previously sequenced and annotated [49] and the draft assembly deposited in the European Nucleotide Archive (EMBL-EBI accession number: CONTIGS: CCBE010000001–CCBE010000111; SCAFFOLDS: HG975453–HG975511). Predictions of subcellular localization of encoded proteins were done in this work using SurfG+ [28], a software dedicated to prediction of potentially surface exposed proteins (PSE) in Gram-positive bacteria. SurfG+ integrates numerous bioinformatics prediction to classify proteins according to their sub-cellular localization: (i) HMM search, to search for a number of motifs that have been described as cell wall anchoring or binding domains (e.g. LPxTG motifs and LysM domains, S-layer homology domain); (ii) LipoP, to identify lipoproteins; (iii) SignalP, to identify proteins that are secreted by the Sec pathway; and (iv) TMMOD, to identify membrane spanning protein segments (TMH) and predict membrane topology. The results from those predictions are then combined to predict localization of proteins: membrane, cytoplasmic, PSE or secreted.
2.3. Whole-cell protein extracts
Whole cell SDS extracts were prepared according to a procedure modified from one previously described [15]. Briefly, 100 mL of stationary phase culture (see above) was harvested by centrifugation (6000 ×g, 10 min, 4 °C) and washed in an equal volume of PBS prior to resuspension in SDS lysis buffer (50 mM Tris–HCl [pH 7.5], 0.3% SDS, 200 mM DTT) to a final OD650 of 20. After 3 freeze/thaw cycles, bacteria were broken by sonication using a Vibra Cell sonicator (Bioblock Scientific, Illkirch, France) equipped with a tapered microtip (4 bursts of 1 min at 1 min intervals, output 2.5). Insoluble materials were removed by centrifugation (10,000 ×g, 10 min, room temperature). The resulting whole-cell protein SDS extract was used for proteomic investigations. This procedure was applied in 3 replicas, on 3 independent cultures, leading to identical results.
2.4. Extraction of surface proteins non-covalently bound to the cell wall using guanidine hydrochloride
Surface layer proteins were extracted according to a procedure modified from one previously described [50]. 100 mL of stationary phase culture (see above) was harvested by centrifugation (6000 ×g, 10 min, 4 °C) and washed in an equal volume of PBS prior to resuspension in 5 M guanidine hydrochloride to a final OD650 of 20. The suspension was incubated 15 min at 50 °C prior to centrifugation (21,000 ×g, 20 min, 30 °C) to eliminate cells. The supernatant was then dialyzed exhaustively against 0.1% SDS in distilled water during 24 h at 4 °C using a 10,000 kDa cutoff Slide-A-Lyer® Dialysis Cassette (ThermoScientific, Rockford, USA) prior to proteomic investigations. This procedure was applied on 3 independent cultures, leading to the identification of the same proteins. Representative results are shown in Supplemental Table 1.
2.5. One-dimensional polyacrylamide gel electrophoresis (1-DE)
Samples in SDS extracts from whole-cell and surface layer fractions were diluted in SDS sample buffer [51] prior to heat-denaturation (10 min, 95 °C). One-dimensional polyacrylamide gel electrophoresis (12.5%) was conducted according to Laemmli [51] on a Protean II xi Cell (Bio-Rad, Hercules, USA) prior to Coomassie Blue-staining using the Bio-Safe reagent (Bio-Rad). The presence of cytoplasmic proteins was checked by western blotting using a serum directed against methylmalonyl-coenzyme A mutase, a P. freudenreichii cytoplasmic specific enzyme (Fig. 1) as previously described [52].
2.6. Enzymatic shaving of surface proteins
100 mL of stationary phase culture (see above) was harvested by centrifugation (6000 ×g, 10 min, 4 °C) and washed in an equal volume of PBS [pH 8.5] containing 5 mM DTT prior to resuspension in 1/10 volume of the same buffer. Sequencing grade modified trypsin (V5111, Promega, Madison, USA) was dissolved in the same buffer (qsp 0.2 g/L) and added to the bacterial suspension. “Shaving” was performed for 1 h at 37 °C in a 0.5 mL reaction volume containing 5 × 109 bacteria and 4 μg of trypsin, with gentle agitation (180 rpm). Bacteria were removed by centrifugation (8000 ×g, 10 min, 20 °C) and the supernatant filtered (0.2 μm, Nalgene) prior to the addition of 1 μg of trypsin to complete digestion of released peptides (16 h, 37 °C). Trypsin digestion of the supernatant was stopped by adding trifluoroacetic acid to a final concentration of 0.15% (v/v). The supernatants containing peptides were then concentrated in a Speed-Vac concentrator prior to nano-LC–MS/MS analysis. The viability of propionibacteria was monitored by CFU counting throughout the shaving procedure. This procedure was applied in 3 replicas, on 3 independent cultures, leading to the identification of the same proteins. Representative results are shown in Supplemental Table 2.
2.7. LC–MS and nano-LC–MS/MS analyses
The dialyzed guanidine hydrochloride extract was analyzed by reverse phase-HPLC on a Vydac 214TP C4 (5 μm particle
size, 2.1 mm by 150 mm length) connected to an HPLC 1100 (Agilent Technologies, Massy, France). Solvent A (milli-Q water containing 1.06‰ (v/v) trifluoroacetic acid) and solvent B (HPLC grade-acetonitrile:Milli-Q water (80:20, v/v)) mixtures containing 1‰(v/v) trifluoroacetic acid were used. A linear gradient which started with 37% of solvent B and reaching 60% over 37 min was used for protein elution. Protein separation was carried out at 40 °C at a flow rate of 0.250 mL/min. This chromatographic system was fitted to a QSTAR XL (MDS SCIEX, Ontario, Canada) equipped with an ion source (ESI) (Proxeon Biosystems A/S, Odense, Denmark) for protein mass measurement. Eluted proteins were electrosprayed into the mass spectrometer operated in positivemode. A full continuous MS scan was carried out and data were collected in the selected mass range of 800 to 3000 m/z. The instrument was calibrated by multipoint calibration using fragment ions that resulted from the collision-induced decomposition of a peptide from β-casein, β-CN (193–209). Data were collected and processed using Analyst QS 1.1 Sciex software and the deconvolution of spectra was carried out using Bioanalyst 1.1.5
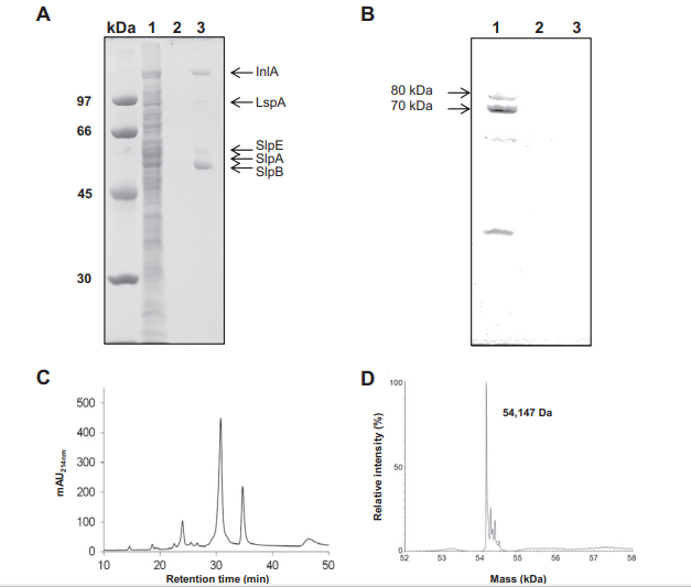
Fig. 1 – Selective extraction of Propionibacterium freudenreichii ITG P20 surface proteins using guanidine hydrochloride. P. freudenreichii was grown in cow's milk ultrafiltrate and harvested in an early stationary phase. A whole-cell protein extract (line 1), the culture supernatant (line 2) and a guanidine hydrochloride extract (line 3) were separated on 10% SDS PAGE. Gels were either Coomassie Blue-stained (A) or transferred onto a PVDF membrane prior to immunolabeling using a rabbit serum directed against the cytoplasmic methylmalonyl-CoA mutase (B). Proteins identified by nano-LC–MS/MS in the guanidine extract (see Table 1 for details) are indicated on line 3 (A). Immunoreactive protein subunits 70,000 Da and 80,000 Da of methylmalonyl-CoA mutase are indicated on line 1 (B). No bacterial lysis is revealed by this western blot analysis. (C) Reverse phase nano-LC profile of the extract (mAU214nm, milliabsorbance units at 214 nm). (D) ESI-MS analysis of the major slpB S-layer protein, indicating an average mass value of 54,147 Da after deconvolution.
For trypsinolyzed proteins, nano-LC experiments were performed using an on-line liquid chromatography tandem mass spectrometry (MS/MS) setup using a Dionex U3000-RSLC nano-LC system fitted to a QSTAR XL (MDS SCIEX, Ontario, Canada) equipped with a nano-electrospray ion source (ESI) (Proxeon Biosystems A/S, Odense, Denmark). Samples were first concentrated on a PepMap 100 reverse-phase column (C18, 5 μm particle size, 300-μm inner diameter (i.d.) by 5 mm length) (Dionex, Amsterdam, The Netherlands). Peptides were separated on a reverse-phase PepMap 100 column (C18, 3 μm particle size, 75 μm i.d. by 150 mm length) (Dionex) at 35 °C, using solvent A (2% (vol/vol) acetonitrile, 0.08% (vol/vol) formic acid, and 0.01% (vol/vol) TFA in deionized water) and solvent B (95% (vol/vol) acetonitrile, 0.08% (vol/vol) formic acid, and 0.01% (vol/vol) TFA in deionized water). A linear gradient from 10 to 50% of solvent B in 40 min was applied for the elution at a flow rate of 0.3 μL/min. Eluted peptides were directly electrosprayed into the mass spectrometer operated in positive mode. A full continuous MS scan was carried out followed by three data-dependent MS/MS scans. Spectra were collected in the selected mass range 400 to 2000 m/z for MS and 60 to 2000 m/z for MS/MS spectra. The three most intense ions from the MS scan were selected individually for collision-induced dissociation (1+ to 4+ charged ions were considered for the MS/MS analysis). The mass spectrometer was operated in data-dependent mode automatically switching between MS and MS/MS acquisition using Analyst QS 1.1 software. The instrument was calibrated by multipoint calibration using fragment ions that resulted from the collisioninduced decomposition of a peptide from β-casein, β-CN (193– 209). The proteins present in the samples were identified from MS and MS/MS data by using MASCOT v. 2.2 software for search into two concatenated databases: (i) a homemade database containing all the predicted proteins of the P. freudenreichii strain CIRM-BIA 129 used in this study and (ii) a portion of the UniProtKB database corresponding to P. freudenreichii. Preliminary experiments, before sequencing of the CIRM-BIA 129 genome, using the UniProtKB database, failed to identify and differentiate the proteins analyzed in this study. Search parameters were set as follows. A trypsin enzyme cleavage was used, the peptide mass tolerance was set to 0.2 Da for both MS and MS/MS spectra, and two variable modifications (oxidation of methionine and deamidation of asparagine and glutamine residues) were selected. For each protein identified in nano-LC–ESI-MS/MS, a minimum of two peptides with a MASCOT score corresponding to a P value below 0.05 were necessary for validation with a high degree of confidence. For automatic validation of the peptides from MASCOT search results, the 1.19.2 version of the IRMa software was used [53].
2.8. In situ surface labeling
The surface labeling procedure was adapted from HagnerMcWhirter et al. [54]. Bacteria were grown and harvested as described above and washed in an equal volume of ice-cold PBS containing 33 mM Tris–HCl [pH 8.5] prior to centrifugation. Bacteria were resuspended in 1/10 volume of ice-cold labeling buffer (PBS containing, 1 M urea, 33 mM Tris–HCl [pH 8.5]). Labeling was performed on ice, in the dark, in a 1 mL reaction volume containing 1010 live and intact bacteria, and 200 pmol of CyDye DIGE Fluor Cy5 minimal dye (GE Healthcare, Orsay, France). Labeling was stopped by adding 1 μmol of Lysine to quench the dye. Labeled bacteria were centrifuged and washed in PBS (pH 7.4), centrifuged and resuspended in SDS lysis buffer (50 mM Tris–HCl [pH 7.5], 0.3% SDS, 200 mM DTT) prior to whole cell protein extraction as described above. Three independent labeling experiments were performed on independent cultures (biological replicas).
2.9. Two-dimensional imaging and spot picking
Whole-cell protein SDS extracts of labeled bacteria were precipitated using the 2D Clean-Up Kit (GE Healthcare) prior to dissolution in destreak rehydration solution (100 μl per sample) added with 2% (w/v) ampholyte containing buffer (IPG-Buffer 4–7, GE Healthcare). Isoelectric focusing was carried out using pH 4 to 7, 18 cm, Immobiline Dry Strips on a Multiphor II electrophoresis system (GE Healthcare) for a total of 60 kVh using a standard procedure described previously [55]. The second dimensional separation was performed on the Ettan™ DALTtwelve electrophoresis system (GE Healthcare) using 14% acrylamide separating gels without a stacking gel at a voltage of 50 V for 1 h and 180 V for about 7 h. Fluorescent images of the gels were immediately acquired on a Typhoon PhosphorImager (GE Healthcare) using the appropriate laser excitation for Cy5 fluorescence. Gels were then fixed and Coomassie Blue-stained as described above. Visible images were acquired on an ImageScanner III (GE Healthcare). Images were further analyzed using Image-Master 2D software. Fluorescent profiles of 2-DE-separated proteins were reproducible in at least three individual experiments. Fluorescent and Coomassie-Blue visible images of the 2D electrophoresis gels were matched to detect surface-exposed proteins. Fluorescent spots corresponding to surface-exposed proteins were excised from 2-DE gels as previously described [15] when detectable by Coomassie-Blue staining. Proteins were identified by tandem mass spectrometry (MS/MS) after an in-gel trypsin digestion adapted from Shevchenko [56]. Briefly, gel pieces were washed with acetonitrile and ammonium bicarbonate solution, and then dried under vacuum in a Speed-Vac concentrator (SVC100H-200; Savant, Thermo Fisher Scientific, Waltham, MA, USA). In-gel trypsin digestion was performed overnight at 37 °C and stopped with spectrophotometric-grade trifluoroacetic acid (TFA) (SigmaAldrich). The supernatants containing peptides were then vacuum dried in a Speed-Vac concentrator and stored at −20 °C until mass spectrometry analysis. Nano-LC–MS/MS analysis was as described above. Three 2D gels were run for each labeling experiment (technical replicas).
2.10. PBMC isolation and induction of cytokine release
Peripheral blood mononuclear cells were isolated from the blood of three healthy donors and reference bacterial strains
were prepared as previously described [57]. Propionibacteria were harvested from fermented milk ultrafiltrate [47] and were either guanidine-extracted (as described above) or left untreated. Propionibacteria, extracted or not, were washed in PBS and resuspended in PBS containing 20% glycerol at the JOURNAL OF PROTEOMICS 112 (2015) 447 – 461 451 same density (turbidimetry Mc Farland unit 3, as previously described). They were then added to PBMCs at a propionibacteriato-immune cell ratio of 5. Finally, a P. freudenreichii guanidine hydrochloride surface protein extract (see above) was extensively dialyzed against PBS, and proteins were quantified using the Bradford assay. Different amounts (0.5 to 50 μg, see Fig. 2) of extracted surface proteins were then added to PBMCs.
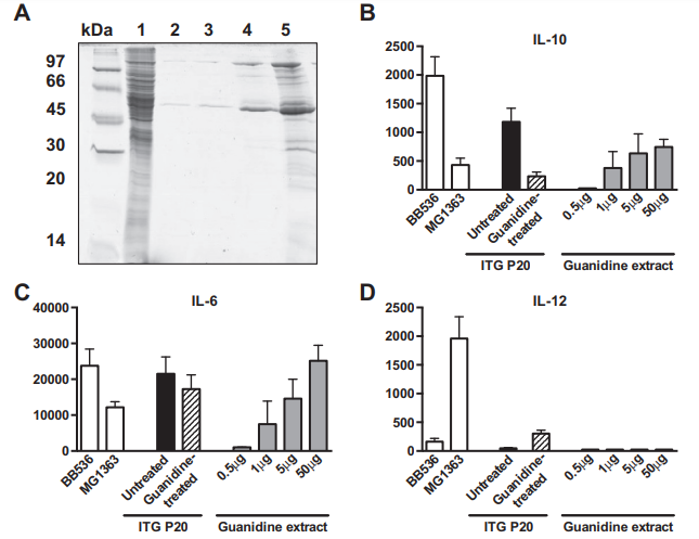
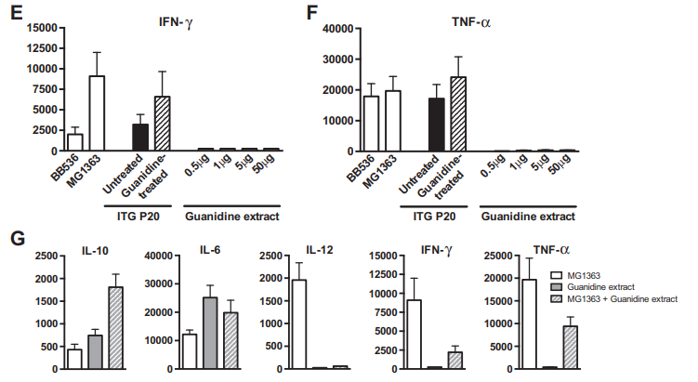
Fig. 2 – Immunomodulatory properties of Propionibacterium freudenreichii ITG P20 surface proteins extracted by guanidine hydrochloride. P. freudenreichii was grown in cow's milk ultrafiltrate and harvested in an early stationary phase. A preparative guanidine hydrochloride extract was made and extensively dialyzed against PBS prior to 12% SDS PAGE electrophoretic analysis (panel A). Samples were a whole-cell protein extract (line 1) and 0.5, 1, 5 and 50 μg of extracted proteins (lines 2 to 5). Panels 2 B to F: production of cytokines by human peripheral blood mononuclear cells (PMMCs) in response to bacteria and/or protein extract. IL-10 (B), IL-6 (C), IL-12 (D), IFN-γ (E) and TNF-α (F) were assessed by ELISA in the supernatants collected from 24 h cultures of human PBMCs stimulated with reference bacteria Bifidobacterium longum BB536 and Lactococcus lactis MG1363 (white bars), with Propionibacterium freudenreichii ITG P20, either untreated (black bars) or guanidine-treated (hatched bars), or with 0.5, 1.0, 5.0 and 50 μg of guanidine-extracted proteins (gray bars). G: production of cytokines by human PBMCs in response to L. lactis MG1363 (white bars), 50 μg of extracted proteins (gray bars), or the combination thereof (hatched bars). Data are expressed in pg/ml as mean ± SEM (n = 3 healthy donors).
After 24-h stimulation, culture supernatants were collected, clarified by centrifugation and stored at −20 °C until cytokine analysis. These were quantified by ELISA as previously described [57] using antibodies provided by R&D systems (Minneapolis, USA) for IL-6 and TNF-α or by BD Pharmingen (BD Biosciences, San Jose, USA) for IL-10, IL-12 and IFN-γ.
3. Results
3.1. Prediction of subcellular localization of the predicted proteins
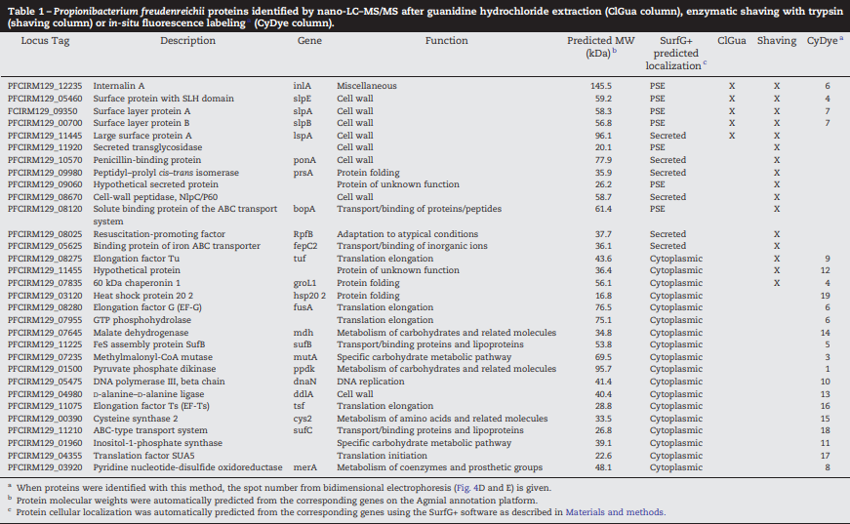
Analysis of the draft genomic sequence of the P. freudenreichii ITG P20 strain revealed 2324 predicted protein-coding genes. This number is close to that in the type strain CIRM-BIA 1T, the first publically available sequenced genome of P. freudenreichii [17], which contained 2439 protein-coding genes. The predicted proteome of ITG P20 has been analyzed with the software SurfG+, dedicated to the prediction of the localization of proteins [28]. This in silico analysis predicted that 1702 proteins were cytoplasmic (73%), 397 were membrane (17%), 59 were secreted (2%) and 180 were potentially surface-exposed (PSE) (8%). Among PSE predicted proteins, 74 are predicted to expose a C-terminal end to the surface, 35 an N-terminal end, 36 are predicted to be lipoproteins, 12 to expose a loop, 8 to exhibit a motif related to cell-wall anchoring or binding domains and one to exhibit a specific motif and a C-terminal end to the surface. 1579 proteins were predicted in the pI ranges 4–7 (68% of the genome) and are thus susceptible to be visible on the 2D gels performed in this study.
3.2. Extraction and analysis of surface proteins non-covalently associated with the cell wall
Surface proteins non-covalently linked to the P. freudenreichii cell wall were extracted using the chaotrope guanidine hydrochloride as previously developed for dairy propionibacteria and described in the Materials and methods section. This guanidine extract was compared to whole-cell SDS extract (Fig. 1A). No main band was detected in the whole-cell SDS extract of P. freudenreichii ITG P20 (lane 1). As a control, no protein was detected in the culture supernatant (lane 2). Accordingly, western blot detection of the cytoplasmic marker enzyme Methylmalonyl-CoA mutase, previously developed to detect P. freudenreichii cell lysis [52], revealed the presence of this marker only in the whole-cell protein extract, yet its absence in the culture supernatant and in the guanidine extract (Fig. 1B).
This suggested that no significant cell lysis occurred, either during growth, or during harvesting of cells. This electrophoretic analysis further revealed the presence of five protein bands in the guanidine extract (lane 3). The gel lane number 3 was sliced and all the strips were subjected to in-gel trypsin digestion followed by nano-LC–MS/MS analysis. Five proteins, listed in Table 1 and indicated in the corresponding gel zones in Fig. 1A, were clearly identified by MS/MS with 3 to 34 unique peptides. These were internalin A (InlA), large surface protein A (lspA), surface protein
with SLH Domain E (slpE), and surface layer protein slpA and slpB. See Supplemental Table 1 for MS/MS details.
The major surface layer protein, SlpB, was further characterized using LC–MS for accurate molecular mass determination. It was separated by reverse phase chromatography and the major peak (elution time 31 min, Fig. 1C) gave a clear MS signal. The corresponding raw MS spectrum (Supplemental Fig. 1) showing a single protein charge state envelope allowed reconstruction of a deconvoluted mass spectrum (Fig. 1D). The deduced average molecular weight of this protein was 54,147 Da, with an accuracy of ±5 Da, considering the thirty most intense charge states of the protein visible on the mass spectrum. This mass did not correspond with any of the ones predicted for the 5 proteins identified in this extract (Table 1).
However, a 29 residue long signal peptide was predicted using the Phobius tool (Stockholm Bioinformatics Centre) in the 556 residue slpB gene sequence (see in Fig. 5). The resulting 527 residue processed protein had a theoretical mass of 54,145 Da, which is compatible with the 54,147 Da experimental mass, considering the accuracy of the spectrometric measure. This confirms that processed slpB is the main protein in the guanidine extract of P. freudenreichii ITG P20.
3.3. Immunomodulatory properties of surface proteins non-covalently associated to the cell wall
A guanidine hydrochloride preparative extraction was performed on P. freudenreichii ITG P20. The immunomodulatory properties of the extract were evaluated on human PBMCs, in comparison with intact propionibacteria. As shown in Fig. 2, the guanidine surface protein extract induced the release of IL-10 and IL-6, in a dose-dependent manner (Fig. 2B & C), with little or no effect on IL-12, TNF-α and IFN-γ (Fig. 2D to F), in human PBMCs. As a comparison, intact P. freudenreichii ITG P20 cells induced release of the 4 cytokines, IL-10, IL-6, TNF-α and IFN-γ. However, guanidine-treated P. freudenreichii ITG P20 lost the ability to induce IL-10. This indicates that the surface extractable proteins trigger the release of the immunomodulatory cytokines IL-10 and IL-6. As a control, same amounts of bovine serum albumin were tested and induced no cytokine secretion in PBMCs (data not shown). In a second experiment, PBMCs were stimulated by the pro-inflammatory L. lactis MG1363, by the guanidine P. freudenreichii surface protein extract, or by the combination thereof (Fig. 2G). L. lactis induced secretion of the proinflammatory cytokines IL-12, IFN-γ and TNF-α. By contrast, the guanidine extract induced IL-10 and IL-6 secretion.
Moreover, this extract, when applied in conjunction with the pro-inflammatory L. lactis, drastically reduced induction of the pro-inflammatory cytokines IL-12, IFN-γ and TNF-α by this bacterium. This confirms the immunomodulatory effect of P. freudenreichii surface proteins, with a marked anti-inflammatory profile.
3.4. Enzymatic shaving and analysis of surface protruding proteins
To go deeply into surface protein characterization, proteins that protrude at the surface of P. freudenreichii cells were subjected to shaving using trypsin acting on live cells. Intact cells were harvested and treated with trypsin for different
time periods. As a first control of the absence of cell lysis, the cellular and extracellular were analyzed by SDS PAGE fractions for different durations of shaving.
No release of intracytoplasmic material was detected, after up to 2 h of shaving (Fig. 3A). Accordingly, the propionibacteria viability was monitored and shown to be constant during this treatment (Fig. 3B). Reverse phase nano-LC analysis of a 1 h shaving supernatant revealed the presence of an eluted material when cells were incubated in the presence of trypsin, yet not in the absence of this enzyme.
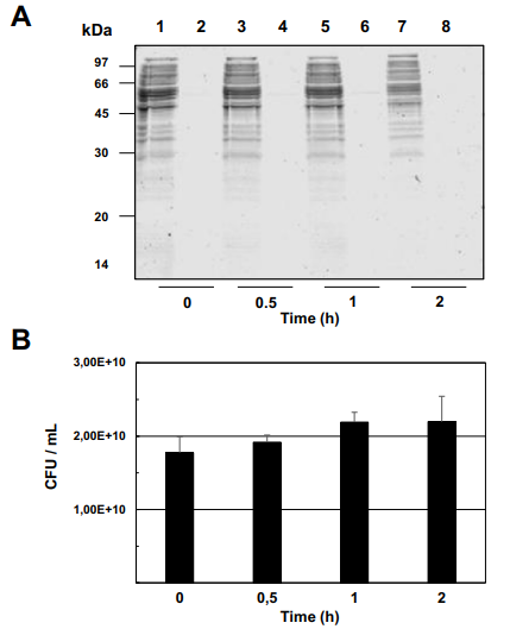
Fig. 3 – Enzymatic shaving of Propionibacterium freudenreichii ITG P20 surface proteins using trypsin. P. freudenreichii was grown in cow's milk ultrafiltrate and harvested in an early stationary phase. Washed cells were subjected to shaving for 0, 0.5, 1 or 2 h, as described in Materials and methods, prior to centrifugation. (A) The resulting pellet (1, 3, 5 and 7) as well as the supernatant (2, 4, 6 and 8) were analyzed by 12% SDS PAGE followed by Coomassie Blue-staining. No bacterial lysis is revealed by the analysis of shaving supernatants, whatever the shaving time. (B) Survival of P. freudenreichii was monitored by CFU counting during shaving and revealed no loss in propionibacterial viability. No bacterial death is revealed during shaving. (C) Surface proteins identified by shaving. After 1 h of shaving with trypsin, the supernatant was isolated and the generated peptides were identified by nano LC–MS/MS.
MS/MS analysis of the eluted peptides led to the identification of 16 unique proteins. As a confirming first result, the 5 proteins identified by guanidine extraction (see above) were also found by shaving. A set of 11 new proteins were also identified here, as shown in Table 1. This includes enzymes involved in cell wall metabolism and the BopA adhesin already described in bifidobacteria [22,23]. See Fig. 3C for MS/MS details. 3.5. Fluorescent labeling and analysis of surface proteins using CyDye DIGE minimal dye To confirm surface accessibility of proteins, intact live cells of P. freudenreichii were subjected to in situ CyDye labeling without loss of viability (data not shown). The resulting cells exhibited intense fluorescence as evidenced by epifluorescence microscopy (Fig. 4A). As a control, no protein was detected by SDS PAGE followed by Coomassie-Blue staining in the labeling reaction (Fig. 4 B), while only one fluorescent band was detected in this fraction (Fig. 4C). By contrast, the bulk majority of fluorescent proteins were observed in the cellular fraction. This was thus separated by two dimensional electrophoresis followed by fluorescence imaging (Fig. 4D) and Coomassie-Blue staining (Fig. 4E). Only fluorescent spots matching with Coomassie-Blue stained ones could be picked for proteomic analysis, while a subset of the fluorescent ones escaped such analysis. As a
control of efficacy, the surface extractible proteins internalin A and the surface layer proteins E, A and B were detected this way. A subset of 15 additional proteins was identified (see Table 1). Four proteins extracted by CyDye were predicted as PSE by SurfG+: SlpA, SlpB, SlpE and InlA. They all belong to protein anchored to the peptidoglycane by an SLH domain and were detected here by the 3 methods, extraction, shaving and labeling. See Supplemental Table 3 for MS/MS details.
3.6. Focus on surface accessibility of two key proteins
Among the surface proteins identified here, we focused on key proteins most likely to play a role in bacterium/host interactions, i.e. adhesion and immunomodulation. We used the peptides detected by shaving (see above) to specify the surface topology of these proteins and identify the exposed domains, most likely to interact with the host. BopA is a bacterial lipoprotein involved in adhesion to epithelial cells in bifidobacteria [58]. Its counterpart in P. freudenreichii exhibited surface exposure and accessibility of its C-terminal part (Fig. 5A). By contrast, the N-terminal's first 103 residues was not detected here. This comprises the 21 residue cleavable signal sequence recognized by the dedicated lipoprotein signal peptidase, as well as the following 82 residues. These are probably embedded in the peptidoglycan thick layer, as they follow the N-terminal lipid-modified cysteine which is tethered to the outer face of the cytoplasmic membrane. This is consistent with their lack of accessibility. S-layer proteins are reportedly involved in adhesion and immunomodulation in other bacteria [24,30,31,59]. They are anchored to the cell wall via electrostatic interactions involving SLH domains and pyruvylated cell wall polymers [31]. As indicated in Fig. 5B, extracellular released peptides covered 49% of P. freudenreichii slpB protein, showing great surface accessibility of the N-terminal part, except for the
cleavable signal sequence. By contrast, the C-terminal region, containing 3 predicted SLH domains, was poorly represented. This confirms the hypothesis that slpB SLH domains are embedded in the peptidoglycan thick layer, thus not accessible to the enzyme.
4. Discussion
4.1. A limited number of proteins validated as surface-exposed
The surface proteome of P. freudenreichii ITG P20 was characterized using three approaches, leading to the identification of a total of 31 different proteins. None of the 3 methods spotted all of them, showing the relevance of using a combination of methods. This number is relatively low, compared to other reports [60]. Among these 31 proteins, 4, i.e. InlA, SlpE, SlpA and SlpB, were confirmed by the 3 methods, 4 additional proteins were confirmed by 2 different methods, while a set of 23 proteins were identified only by one method. S-layer proteins, previously thought to be composed of a single protein molecular species, are more complex in Gram-positive bacteria such as L. acidophilus, the S-layer of which contains 37 proteins [61], in addition to the previously described SlpA. In P. freudenreichii ITG P20, at least 5 proteins are extracted using guanidine, SlpB being the major one, with a MS-measured mass of 54,147 Da, which is close to the 54,145 Da size predicted for SlpB after cleavage of the signal peptide (this work, predicted from the slpB gene).
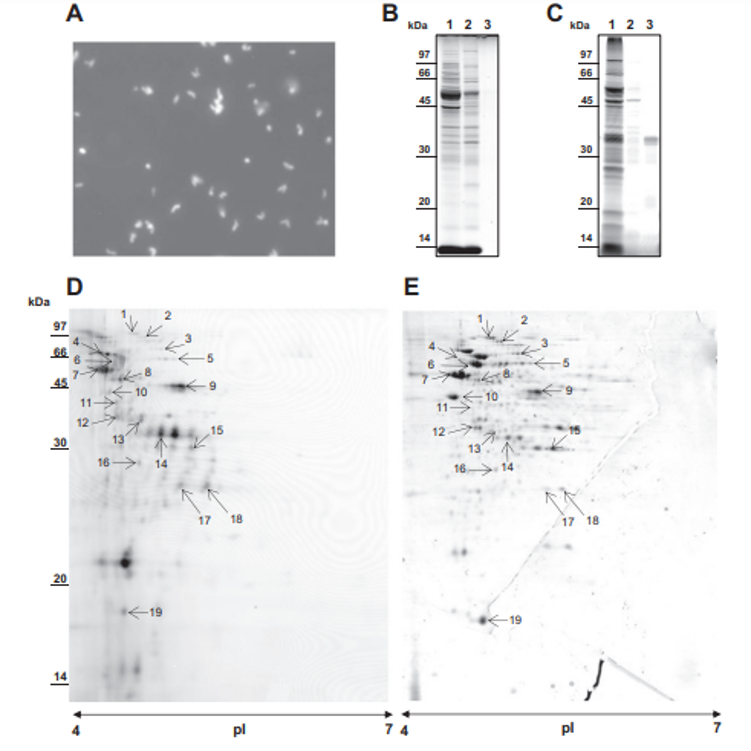
Fig. 4 – Selective in situ labeling of Propionibacterium freudenreichii ITG P20 surface proteins using CyDye DIGE Fluor Cy5 minimal dye. P. freudenreichii was grown in cow's milk ultrafiltrate and harvested in an early stationary phase. Washed cells were subjected to CyDye surface labeling as described in Materials and methods prior to examination under a fluorescence microscope (A). (B) SDS PAGE analysis of labeled P. freudenreichii whole-cell protein SDS extract (1), of labeled P. freudenreichii lysozyme extract (2) and of the labeling reaction supernatant, after centrifugation of bacteria (3). (C) Fluorescence imaging, using a Typhoon PhosphorImager, of the same gel. Fluorescent proteins were mainly detected in the whole-cell protein extract, while only one diffused into the extracellular medium. (D) The CyDye-labeled P. freudenreichii whole-cell protein extract was further separated by two-dimensional gel electrophoresis prior to fluorescence imaging on a Typhoon PhosphorImager. (E) The same gel was Coomassie Blue-stained prior to scanning on an ImageScanner III. Coomassie Blue-stained spots co-localized with fluorescent ones are indicated by an arrow.
Enzymatic shaving of Gram-positive bacteria can lead to the identification of up to 72 proteins, of which 6 to 70% are predicted as cytoplasmic, depending on the report [60]. Here, we describe 16 distinct P. freudenreichii proteins, with only 3 (18%) predicted as cytoplasmic, including Grol1 and Tuf, known as moonlighting surface exposed proteins in other Gram-positive bacteria [36,38,62–65].
Surface labeling with a NHS-ester coupled CyDye confirmed surface exposure of P. freudenreichii proteins. This method, first developed for eukaryotic cells, was applied to decipher M. genitalium surfaceome, allowing the identification of 61 distinct proteins [40]. To our knowledge, this work constitutes the first application of this labeling to a food bacterium. It identified 22 proteins in P. freudenreichii. Among these, 7 were also identified by extraction and/or shaving, while shaving revealed 9 proteins not spotted by labeling. This is probably due to the limits of 2D electrophoretic techniques, as surface proteins may be insoluble, out of the pH range, or underrepresented. Surprisingly, a set of 15 proteins (Table 1, bottom lines, all predicted as cytoplasmic) were detected only by labeling. These should be considered with caution, as CyDyes are small molecules able to diffuse through the peptidoglycan cell wall, and thus reaching the cell membrane, while the trypsin enzyme does not.
4.2. Proteins with peptidoglycan binding domain
Proteins containing SLH domains are typical surface exposed proteins. Four were identified in this study: SlpA, B and E and InlA. SLH domains, illustrated in Fig. 5, are involved in the interaction of such proteins, the so-called S-layer associated proteins (SLAPs) [61], with cell wall polymers and act as anchorage structures in the Gram positive cell wall. According to bioinformatic prediction, nine proteins present SLH domains in the draft genome of P. freudenreichii ITG P20. Five of them have neither been extracted, nor identified by surface proteomic analysis. They may not be expressed in our growth conditions, or their secondary structure and cell wall-anchoring may prevent these proteins from protruding at the cell surface. Another PSE protein (PFCIRM129_11920), annotated as a secreted transglycosidase, possesses N-terminal signal peptides (18 residues) and a peptidoglycan binding domain LysM. This cell wall associated protein is homologous to a Streptomyces sviceus peptidoglycan binding protein involved in a cell wall macromolecule catabolic process (Genbank ID CM000951).
4.3. Lipoproteins
Two proteins presented a lipoprotein domain. The first one, PFCIRM129_09060, a putative uncharacterized protein also found in other actinobacteria including Mobiluncus, Bifidobacterium and Arthrobacter, displays a lipoprotein domain and an “Excalibur” domain (extracellular calcium-binding region). The second one was identified as BopA, a protein belonging to the ABC superfamily with an ATP binding cassette, with extensive homology with its counterpart in bifidobacteria. In P. freudenreichii ITG P20, it displays a 32 residue N-terminal peptide, a large bacterial extracellular solute binding domain (107 to 475) and a lipoprotein domain (prokar prediction). As none of them possesses a membrane spanning protein segment, C-terminal parts of these proteins probably protrude out of the surface of the cell. This is in accordance with the position of the peptides identified by shaving along the BopA protein (Fig. 5).
4.4. Proteins predicted to be secreted
A subset of 6 proteins was predicted as secreted, mainly because of the presence of a signal peptide. However, they match with enzymes known to play a role within the cell wall. The protein PonA (PFCIRM129_10570), as an example, belongs to a family of bifunctional transglycosidase/transpeptidases involved in the polymerization of murein glycan chains, a key step in the synthesis of a cell wall peptidoglycan. The cell wall peptidase NlpC/P60 (PFCIRM129_08670) is a conserved protein found on the surface of several bacteria. Its homologue YgjB is involved in peptidoglycan hydrolysis in L. lactis [66]. The peptidyl–prolyl cis–trans isomerase (PFCIRM129_09980) is thought to be involved in protein folding. Its counterpart (PrsA) is involved in protein post-export processes in other Gram positive bacteria [67]. Acting as a cell wall foldase, it determines key surface properties.
 Fig. 5 – Aminoacid sequence predicted for the BopA (panel A) and SlpB (panel B) proteins and surface accessibility. The signal sequences are underlined. The potent trypsin cleavage sites are indicated by dark triangles following lysine (K) and arginine residues (R). The peptides detected in the shaving extracellular fraction are highlighted, evidencing the surface-accessible cleavage sites. The 3 SLH domains detected in slpB are indicated.
Fig. 5 – Aminoacid sequence predicted for the BopA (panel A) and SlpB (panel B) proteins and surface accessibility. The signal sequences are underlined. The potent trypsin cleavage sites are indicated by dark triangles following lysine (K) and arginine residues (R). The peptides detected in the shaving extracellular fraction are highlighted, evidencing the surface-accessible cleavage sites. The 3 SLH domains detected in slpB are indicated.
4.5. Proteins predicted to be cytoplasmic
Among the 31 proteins experimentally identified as surface exposed, 18 are predicted by SurfG+ to be cytoplasmic. At least 6 of these 18 proteins have already been reported to be localized at the surface of other bacteria: elongation factors Tu, Ts and G; heat shock protein 20, 60 kDa chaperonin, and pyruvate kinase. Proteins able to exhibit distinct biological functions in relation with distinct subcellular localization are named “moonlighting” proteins [68,69]. Glycolytic enzymes, chaperones and translation factors are frequently reported to be found at the surfaces of several bacterial species with a surface-specific role in adhesion, plasminogen-binding or modulation of the host immune response in both pathogenic and probiotic bacteria [69].
Mechanisms involved in the exportation of these proteins are still not elucidated. Their secretion, previously thought to
be due to cell lysis, is in fact tightly regulated, signal-peptide independent, not coupled to translation, and occurs in response to specific stimuli and in stationary phase [70]. Both Gram-negative and Gram-positive may also produce membrane vesicles allowing exportation of moonlighting proteins [71]. Moonlighting proteins, lacking a well-known and characterized export signal, are predicted as cytoplasmic by bio-informatics tools such as SurfG+. However, new targeting signals are being identified in such proteins [72].
4.6. Surface proteins involved in Propionibacterium/host interactions
Internalin A (InlA) is known to be involved in adhesion in several bacteria including the pathogen Listeria monocytogenes. This adhesin was also found in the lactic acid bacteria Carnobacterium maltaromaticum and Lactobacillus plantarum [73]. Its sequence contains several LRRs (leucine rich repeats) known to participate in protein/protein interactions and found in various cell adhesion molecules. In bifidobacteria, BopA was shown to be involved in adhesion to human epithelial cells [58,74]. Interestingly, BopA expression level is linked with adhesion and with anti-inflammatory properties in Bifidobacterium bifidum. In P. freudenreichii, InlA and BopA are thus probably involved in interaction with the host digestive tract, considering that adhesive properties were described for this probiotic bacterium [18–20,43]. Moonlighting proteins on the surface of the bacteria may coincide with a new function,
including interaction with the host [69]. This was described for the elongation factor EF-Tu which, when surface exposed, acts as a host-induced adhesin [65,75] and which was identified in P. freudenreichii ITG P20 in this work.
This work also pointed out the role of P. freudenreichii ITG P20 surface proteins in immunomodulation. In several strains of P. freudenreichii, we have previously shown that the removal of S-layer associated proteins (SLAPs) by guanidine extraction leads to drastic modifications of immunomodulatory properties, including abolishment of the ability to trigger release of the regulatory cytokine IL-10 [13]. The ITG P20 strain was further identified as the most efficient inducer of IL-10 [12]. In the present study, we show that a mixture of SLAPs, extracted from the P. freudenreichii ITG P20 surface, is at least partly responsible for the induction of the regulatory cytokines Il-10 and Il-6. This includes InlA, LspA, SlpE, SlpA and SlpB, but the precise role of each of these remains to be confirmed at the molecular level. No common function shared by all proteins with an S-layer domain in bacteria has been found up to now. However, they were shown to play a central role in interactions of several probiotic bacteria with the host. Indeed, they may be involved in tolerance to the digestive stresses, adhesion properties [76] or immunomodulatory properties of these bacteria. As an example, the main S-layer protein is involved in the modulation of immune and epithelial cells by L. acidophilus NCFM [30] and L. helveticus [77]. The anti-inflammatory response to surface proteins of bacteria used as a probiotic or as a fermentation starter is studied thoroughly throughout the world and no general rule, applicable to all bacteria and their surface proteins, is evidenced. This work thus opens new perspectives for the use of selected dairy propionibacteria strains as probiotics for specific populations.
5. Conclusion
As a conclusion, a combination of three different methods was used to inventory the surface proteins of P. freudenreichii. Proteins with different predicted localizations were detected. A subset of surface proteins is involved in the structure, functions and metabolism of the cell wall (transglycosidase, transpeptidase, peptidase, D-Ala–D-Ala ligase, S-layer type proteins), as expected in such an approach. Accordingly, others are involved in binding and transport of extracellular solutes (solute binding protein, iron transport). Some proteins were already described in other bacteria as involved in complex interactions between the bacterium and the host. This includes the conserved adhesin InlA and the lipoprotein BopA involved in adhesion and immunomodulation in bifidobacteria. Some of the detected moonlighting proteins are conserved and were previously shown to play a role in bacterium/host interactions, including adhesion and immune response: EF-Tu, EF-Ts, GroEL, as well as 3 distinct S-layer type proteins. This first inventory of P. freudenreichii surface proteins constitutes the basis for the elucidation of the mechanisms involved in its interaction with its environment. We show, for the first time, the role of P. freudenreichii surface proteins in cytokine induction. The detected genes are presently candidates for gene overexpression and inactivation, an approach which will confirm their physiological and/or functional role.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jprot.2014.07.018.
Acknowledgments
This work was part of the “SURFING” project, Starter SURFace against INflammation of the Gut, ANR-2010-ALIA-016, financed by the French National Agency for Research. GJ thanks the CRITT Santé Bretagne for assistance in writing the ANR Grant. The authors thank “Actalia Dairy Products” for providing the ITG P20 strain.
REFERENCES
[1] FAO/WHO. Guidelines for the evaluation of probiotics in food. Geneva, Switz: Food and Agriculture Organization of the United Nations and World Health Organization Working Group report; 2002.
[2] Bouglé D, Roland N, Lebeurrier F, Arhan P. Effect of propionibacteria supplementation on fecal bifidobacteria and segmental colonic transit time in healthy human subjects. Scand J Gastroenterol Feb 1999;34(2):144–8.
[3] Roland N, Bougle D, Lebeurrier F, Arhan P, Maubois JL. Propionibacterium freudenreichii stimulates the growth of Bifidobacterium bifidum in vitro and increases fecal bifidobacteria in healthy human volunteers. Int Dairy J 1998; 8(6):587–8.
[4] Hojo K, Yoda N, Tsuchita H, Ohtsu T, Seki K, Taketomo N, et al. Effect of ingested culture of Propionibacterium freudenreichii ET-3 on fecal microflora and stool frequency in healthy females. Biosci Microflora 2002;21(2):115–20.
[5] Kaneko T. A novel bifidogenic growth stimulator produced by Propionibacterium freudenreichii. Biosci Microflora 1999;18(2): 73–80.
[6] Seki K, Nakao H, Umino H, Isshiki H, Yoda N, Tachihara R, et al. Effects of fermented milk whey containing novel bifidogenic growth stimulator produced by Propionibacterium on fecal bacteria, putrefactive metabolite, defecation frequency and fecal properties in senile volunteers needed serious nursing-care taking enteral nutrition by tube feeding. J Intest Microbiol 2004;18(2):107–15.
[7] Ouwehand AC, Lagstrom H, Suomalainen T, Salminen S. Effect of probiotics on constipation, fecal azoreductase activity and fecal mucin content in the elderly. Ann Nutr Metab 2002;46(3–4):159–62.
[8] Mitsuyama K, Masuda J, Yamasaki H, Kuwaki K, Kitazaki S, Koga H, et al. Treatment of ulcerative colitis with milk whey culture with Propionibacterium freudenreichii. J Intest Microbiol 2007;21(2):143–7.
[9] Jan G, Belzacq AS, Haouzi D, Rouault A, Metivier D, Kroemer G, et al. Propionibacteria induce apoptosis of colorectal carcinoma cells via short-chain fatty acids acting on mitochondria. Cell Death Differ Feb 2002;9(2):179–88.
[10] Lan A, Lagadic-Gossmann D, Lemaire C, Brenner C, Jan G. Acidic extracellular pH shifts colorectal cancer cell death from apoptosis to necrosis upon exposure to propionate and acetate, major end-products of the human probiotic
propionibacteria. Apoptosis Dec 29 2007;12:573–91.
[11] Cousin FJ, Jouan-Lanhouet S, Dimanche-Boitrel MT, Corcos L, Jan G. Milk fermented by Propionibacterium freudenreichii induces apoptosis of HGT-1 human gastric cancer cells. PLoS ONE 2012;7(3):e31892.
[12] Foligné B, Breton J, Mater D, Jan G. Tracking the microbiome functionality: focus on Propionibacterium species. Gut Feb 6 2013;62(8):1227–8.
[13] Foligné B, Deutsch SM, Breton J, Cousin FJ, Dewulf J, Samson M, et al. Promising immunomodulatory effects of selected strains of dairy propionibacteria as evidenced in vitro and in vivo. Appl Environ Microbiol Dec 2010;76(24):
8259–64.
[14] Deutsch SM, Parayre S, Bouchoux A, Guyomarc'h F, Dewulf J, Dols-Lafargue M, et al. Contribution of surface beta-glucan polysaccharide to physicochemical and immunomodulatory properties of Propionibacterium freudenreichii. Appl Environ Microbiol Mar 2012;78(6):1765–75.
[15] Leverrier P, Vissers JP, Rouault A, Boyaval P, Jan G. Mass spectrometry proteomic analysis of stress adaptation reveals both common and distinct response pathways in Propionibacterium freudenreichii. Arch Microbiol Mar 2004; 181(3):215–30.
[16] Hervé C, Fondrevez M, Cheron A, Barloy-Hubler F, Jan G. Transcarboxylase mRNA: a marker which evidences P. freudenreichii survival and metabolic activity during its transit in the human gut. Int J Food Microbiol Feb 15 2007; 113(3):303–14.
[17] Falentin H, Deutsch SM, Jan G, Loux V, Thierry A, Parayre S, et al. The complete genome of Propionibacterium freudenreichii CIRM-BIA1, a hardy actinobacterium with food and probiotic applications. PLoS ONE 2010;5(7):e11748.
[18] Ouwehand AC, Tolkko S, Kulmala J, Salminen S, Salminen E. Adhesion of inactivated probiotic strains to intestinal mucus. Lett Appl Microbiol Jul 2000;31(1):82–6.
[19] Thiel A, Eikmanns B, Salminen S, Ouwehand AC. In vitro adhesion of propionibacteria to human intestinal mucus. Ital J Food Sci 2004;16(2):245–53.
[20] Tuomola EM, Ouwehand AC, Salminen SJ. Human ileostomy glycoproteins as a model for small intestinal mucus to investigate adhesion of probiotics. Lett Appl Microbiol Mar 1999;28(3):159–63.
[21] Huang Y, Adams MC. An in vitro model for investigating intestinal adhesion of potential dairy propionibacteria
probiotic strains using cell line C2BBe1. Lett Appl Microbiol 2003;36(4):213–6.
[22] Riedel CU, Foata F, Goldstein DR, Blum S, Eikmanns BJ. Interaction of bifidobacteria with Caco-2 cells-adhesion and impact on expression profiles. Int J Food Microbiol Jul 1 2006; 110(1):62–8.
[23] Preising J, Philippe D, Gleinser M, Wei H, Blum S, Eikmanns BJ, et al. Selection of bifidobacteria based on adhesion and anti-inflammatory capacity in vitro for amelioration of murine colitis. Appl Environ Microbiol 2010;76(9):3048–51.
[24] Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol Mar 2010;8(3):171–84.
[25] van de Guchte M, Chaze T, Jan G, Mistou MY. Properties of probiotic bacteria explored by proteomic approaches. Curr Opin Microbiol 2012;15:1–9.
[26] Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol Jul 16 2004;340(4):783–95.
[27] Schneewind O, Missiakas DM. Protein secretion and surface display in Gram-positive bacteria. Philos Trans R Soc Lond B Biol Sci Apr 19 2012;367(1592):1123–39.
[28] Barinov A, Loux V, Hammani A, Nicolas P, Langella P, Ehrlich D, et al. Prediction of surface exposed proteins in Streptococcus pyogenes, with a potential application to other Gram-positive bacteria. Proteomics Jan 2009;9(1):61–73.
[29] Ashida N, Yanagihara S, Shinoda T, Yamamoto N. Characterization of adhesive molecule with affinity to Caco-2 cells in Lactobacillus acidophilus by proteome analysis. J Biosci Bioeng Oct 2011;112(4):333–7.
[30] Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci U S A Dec 9 2008;105(49):
19474–9.
[31] Mesnage S, Fontaine T, Mignot T, Delepierre M, Mock M, Fouet A. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J Sep 1 2000;19(17):4473–84.
[32] Kern J, Wilton R, Zhang R, Binkowski TA, Joachimiak A, Schneewind O. Structure of surface layer homology (SLH)
domains from Bacillus anthracis surface array protein. J Biol Chem Jul 22 2011;286(29):26042–9.
[33] Rodriguez-Ortega MJ, Norais N, Bensi G, Liberatori S, Capo S, Mora M, et al. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat Biotechnol Feb 2006;24(2): 191–7.
[34] Bensi G, Mora M, Tuscano G, Biagini M, Chiarot E, Bombaci M, et al. Multi high-throughput approach for highly selective identification of vaccine candidates: the Group A Streptococcus case. Mol Cell Proteomics Jun 2012;11(6):M111.
[35] Ythier M, Resch G, Waridel P, Panchaud A, Gfeller A, Majcherczyk P, et al. Proteomic and transcriptomic profiling of Staphylococcus aureus surface LPXTG-proteins: correlation with agr genotypes and adherence phenotypes. Mol Cell Proteomics Nov 2012;11(11):1123–39.
[36] Bohle LA, Riaz T, Egge-Jacobsen W, Skaugen M, Busk OL, Eijsink VG, et al. Identification of surface proteins in Enterococcus faecalis V583. BMC Genomics 2011;12:135.
[37] Michaux C, Romero Saavedra LF, Reffuveille F, Bernay B, Goux D, Hartke A, et al. The cold-shock RNA binding protein CspR is also exposed to the surface of Enterococcus faecalis. Microbiology Aug 16 2013;159:2153–61.
[38] Berlec A, Zadravec P, Jevnikar Z, Strukelj B. Identification of candidate carrier proteins for surface display on Lactococcus lactis by theoretical and experimental analyses of the surface proteome. Appl Environ Microbiol Feb 2011;77(4): 1292–300.
[39] Mayrhofer C, Krieger S, Allmaier G, Kerjaschki D. DIGE compatible labeling of surface proteins on vital cells in vitro and in vivo. Proteomics Jan 2006;6(2):579–85.
[40] Parraga-Nino N, Colome-Calls N, Canals F, Querol E, Ferrer-Navarro M. A comprehensive proteome of Mycoplasma
genitalium. J Proteome Res May 2012;23.
[41] Lopez C, Maillard MB, Briard-Bion V, Camier B, Hannon JA. Lipolysis during ripening of Emmental cheese considering organization of fat and preferential localization of bacteria. J Agric Food Chem Aug 9 2006;54(16):5855–67.
[42] Cousin FJ, Deutsch SM, Perez Chaia A, Foligné B, Jan G. Interactions between probiotic dairy propionibacteria and the intestinal epithelium. Curr Immunol Rev 2012;8(3):216–26.
[43] Ouwehand AC, Suomalainen T, Tolkko S, Salminen S. In vitro adhesion of propionic acid bacteria to human intestinal mucus. Lait 2002;82(1):123–30.
[44] Richoux R, Faivre E, Kerjean JR. Effect of NaCl content on lactate fermentation by Propionibacterium freudenreichii in small scale Swiss-type cheeses. Lait 1998;78(3):319–31.
[45] Thierry A, Richoux R, Kerjean JR. Isovaleric acid is mainly produced by Propionibacterium freudenreichii in Swiss cheese. Int Dairy J 2004;14(9):801–7.
[46] Bryson K, Loux V, Bossy R, Nicolas P, Chaillou S, van de Guchte M, et al. AGMIAL: implementing an annotation strategy for prokaryote genomes as a distributed system. Nucleic Acids Res 2006;34(12):3533–45.
[47] Cousin FJ, Foligne B, Deutsch SM, Massart S, Parayre S, Le Loir Y, et al. Assessment of the probiotic potential of a dairy product fermented by Propionibacterium freudenreichii in piglets. J Agric Food Chem Aug 15 2012;60(32):7917–27.
[48] Malik AC, Reinbold GW, Vedamuthu ER. An evaluation of the taxonomy of Propionibacterium. Can J Microbiol 1968;14:1185–91.
[49] Falentin H, Deutsch SM, Loux V, Hammani A, Buratti J, Parayre S, et al. Permanent draft genome of the probiotic
strain Propionibacterium freudenreichii CIRM-BIA 129 (ITG P20). Stand Genomic Sci 2014 (Submitted for publication).
[50] Lortal S, Rouault A, Cesselin B, Sleytr UB. Paracrystalline surface layers of dairy propionibacteria. Appl Environ Microbiol 1993;59(8):2369–74.
[51] Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature Aug 15
1970;227:680–5.
[52] Valence F, Richoux R, Thierry D, Palva A, Lortal S. Autolysis of Lactobacillus helveticus and Propionibacterium freudenreichii in Swiss cheeses: first evidence by using species-specific lysis markers. J Dairy Res 1998;65(4):609–20.
[53] Dupierris V, Masselon C, Court M, Kieffer-Jaquinod S, Bruley C. A toolbox for validation of mass spectrometry peptides identification and generation of database: IRMa. Bioinformatics Aug 1 2009;25(15):1980–1.
[54] Hagner-McWhirter A, Winkvist M, Bourin S, Marouga R. Selective labeling of cell-surface proteins using CyDye DIGE Fluor minimal dyes. J Vis Exp 2008;21.
[55] Anastasiou R, Leverrier P, Krestas I, Rouault A, Kalantzopoulos G, Boyaval P, et al. Changes in protein synthesis during thermal adaptation of Propionibacterium freudenreichii subsp. shermanii. Int J Food Microbiol Feb 10 2006; 108(3):301–14.
[56] Shevchenko A, Sunyaev S, Liska A, Bork P, Shevchenko A. Nanoelectrospray tandem mass spectrometry and sequence similarity searching for identification of proteins from organisms with unknown genomes. Methods Mol Biol 2003; 211:221–34.
[57] Foligné B, Nutten S, Grangette C, Dennin V, Goudercourt D, Poiret S, et al. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J Gastroenterol Jan 14 2007;13(2):236–43.
[58] Gleinser M, Grimm V, Zhurina D, Yuan J, Riedel CU. Improved adhesive properties of recombinant bifidobacteria expressing the Bifidobacterium bifidum-specific lipoprotein BopA. Microb Cell Fact 2012;11:80.
[59] Lebeer S, Vanderleyden J, De Keersmaecker SC. Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev Dec 2008;72(4):728–64.
[60] Olaya-Abril A, Jimenez-Munguia I, Gomez-Gascon L, Rodriguez-Ortega MJ. Surfomics: shaving live organisms for a fast proteomic identification of surface proteins. J Proteome Apr 26 2014;97:164–76.
[61] Johnson B, Selle K, O'Flaherty S, Goh YJ, Klaenhammer T. Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiology Nov 2013;159(Pt 11):2269–82.
[62] Peng Z, Krey V, Wei H, Tan Q, Vogelmann R, Ehrmann MA, et al. Impact of actin on adhesion and translocation of Enterococcus faecalis. Arch Microbiol Dec 21 2013;196:109–17.
[63] Turpin W, Humblot C, Noordine ML, Thomas M, Guyot JP. Lactobacillaceae and cell adhesion: genomic and functional screening. PLoS ONE 2012;7(5):e38034.
[64] Dhanani AS, Bagchi T. The expression of adhesin EF-Tu in response to mucin and its role in Lactobacillus adhesion and competitive inhibition of enteropathogens to mucin. J Appl Microbiol Aug 2013;115(2):546–54.
[65] Yuan J, Wang B, Sun Z, Bo X, Yuan X, He X, et al. Analysis of host-inducing proteome changes in Bifidobacterium longum NCC2705 grown in vivo. J Proteome Res Jan 2008;7(1):375–85.
[66] Redko Y, Courtin P, Mezange C, Huard C, Chapot-Chartier MP. Lactococcus lactis gene yjgB encodes a gamma-Dglutaminyl-L-lysyl-endopeptidase which hydrolyzes peptidoglycan. Appl Environ Microbiol Sep 2007;73(18): 5825–31.
[67] Guo L, Wu T, Hu W, He X, Sharma S, Webster P, et al. Phenotypic characterization of the foldase homologue PrsA
in Streptococcus mutans. Mol Oral Microbiol Apr 2013;28(2): 154–65.
[68] Henderson B, Martin A. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun Sep 2011; 79(9):3476–91.
[69] Wang G, Xia Y, Cui J, Gu Z, Song Y, Chen YQ, et al. The roles of moonlighting proteins in bacteria. Curr Issues Mol Biol Jul 22 2013;16(2):15–22.
[70] Yang CK, Ewis HE, Zhang X, Lu CD, Hu HJ, Pan Y, et al. Nonclassical protein secretion by Bacillus subtilis in the
stationary phase is not due to cell lysis. J Bacteriol Oct 2011; 193(20):5607–15
[71] Lee J, Lee EY, Kim SH, Kim DK, Park KS, Kim KP, et al. Staphylococcus aureus extracellular vesicles carry biologically active beta-lactamase. Antimicrob Agents Chemother Jun 2013;57(6):2589–95.
[72] Yang CK, Zhang XZ, Lu CD, Tai PC. An internal hydrophobic helical domain of Bacillus subtilis enolase is essential but not sufficient as a non-cleavable signal for its secretion. Biochem Biophys Res Commun Mar 15 2014;446:901–5.
[73] Leisner JJ, Hansen MA, Larsen MH, Hansen L, Ingmer H, Sorensen SJ. The genome sequence of the lactic acid bacterium, Carnobacterium maltaromaticum ATCC 35586 encodes potential virulence factors. Int J Food Microbiol Jan 16 2012;152(3):107–15.
[74] Guglielmetti S, Tamagnini I, Mora D, Minuzzo M, Scarafoni A, Arioli S, et al. Implication of an outer surface lipoprotein in adhesion of Bifidobacterium bifidum to Caco-2 cells. Appl Environ Microbiol Aug 2008;74(15):4695–702.
[75] Balasubramanian S, Kannan TR, Baseman JB. The surface-exposed carboxyl region of Mycoplasma pneumonia elongation factor Tu interacts with fibronectin. Infect Immun Jul 2008;76(7):3116–23.
[76] Liu Z, Shen T, Zhang P, Ma Y, Qin H. Lactobacillus plantarum surface layer adhesive protein protects intestinal epithelial cells against tight junction injury induced by enteropathogenic Escherichia coli. Mol Biol Rep Jun 2011; 38(5):3471–80.
[77] Taverniti V, Stuknyte M, Minuzzo M, Arioli S, De Noni I, Scabiosi C, et al. S-layer protein mediates the stimulatory
effect of Lactobacillus helveticus MIMLh5 on innate immunity. Appl Environ Microbiol Feb 2013;79(4):1221–31.