A Peptidomic Analysis of Human Milk Digestion in the Infant Stomach Reveals Protein-Specific Degradation Patterns
The in vitro digestion of isolated milk proteins results in milk peptides with a variety of actions, including antimicrobial (1), immunomodulatory (2), calcium-binding (3), probiotic (4), and antihypertensive (5) actions.
David C. Dallas,4,5* Andres Guerrero,6 Nora Khaldi,4,7 Robyn Borghese,8 Aashish Bhandari,4 Mark A. Underwood,5,8 Carlito B. Lebrilla,5,6 J. Bruce German,4,5 and Daniela Barile4,5
4Department of Food Science, 5Foods for Health Institute, and 6Department of Chemistry, University of California at Davis, Davis, CA; 7University College Dublin Conway Institute of Biomolecular and Biomedical Research, School of Medicine and Medical Sciences, UCD Complex and Adaptive Systems Laboratory, University College Dublin, Dublin, Republic of Ireland; and 8Department of Pediatrics, University of California at Davis, Sacramento, CA
© 2014 American Society for Nutrition.
Manuscript received November 19, 2013. Initial review completed January 9, 2014. Revision accepted March 17, 2014. 815
First published online April 3, 2014; doi:10.3945/jn.113.185793.
Abstract
In vitro digestion of isolated milk proteins results in milk peptides with a variety of actions. However, it remains unclear to what degree protein degradation occurs in vivo in the infant stomach and whether peptides previously annotated for bioactivity are released. This study combined nanospray LC separation with time-of-flight mass spectrometry, comprehensive structural libraries, and informatics to analyze milk from 3 human mothers and the gastric aspirates from their 4- to 12-d-old postpartum infants. Milk from the mothers contained almost 200 distinct peptides, demonstrating enzymatic degradation of milk proteins beginning either during lactation or between milk collection and feeding. In the gastric samples, 649 milk peptides were identified, demonstrating that digestion continues in the infant stomach. Most peptides in both the intact milk and gastric samples were derived from b-casein. The numbers of peptides from b-casein, lactoferrin, a-lactalbumin, lactadherin, k-casein, serum albumin, bile salt–associated lipase, and xanthine dehydrogenase/oxidase were significantly higher in the gastric samples than in the milk samples (P < 0.05). A total of 603 peptides differed significantly in abundance between milk and gastric samples (P < 0.05). Most of the identified peptides have previously identified biologic activity. Gastric proteolysis occurs in the term infant in the first 2 wk of life, releasing biologically active milk peptides with immunomodulatory and antibacterial properties of clinical relevance to the proximal intestinal tract. Data are available via ProteomeXchange (identifier PXD000688). J. Nutr. 144: 815–820, 2014.
1 Supported in part by the National Science Foundation Graduate Research Fellowship Program (D.C.D.) and a USDA National Institute for Food and Agriculture Postdoctoral Fellowship (D.C.D.) and the NIH (R01 HD059127, UL1 TR000002 and R01AT007079). The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository (19) with the data set identifier PXD000688.
2 Author disclosures: D. C. Dallas, A. Guerrero, N. Khaldi, R. Borghese, A. Bhandari, M. A. Underwood, C. B. Lebrilla, J. B. German, and D. Barile, no conflicts of interest.
3 Supplemental Figures 1–3 are available from the ‘‘Online Supporting Material’’ link in the online posting of the article and from the same link in the online table of contents at http://jn.nutrition.org.
* To whom correspondence should be addressed. E-mail: dcdallas@ucdavis.edu.
Introduction
The in vitro digestion of isolated milk proteins results in milk peptides with a variety of actions, including antimicrobial (1), immunomodulatory (2), calcium-binding (3), probiotic (4), and antihypertensive (5) actions. In many cases, the released fragments express activity different from that of the parent protein. Such findings led to the hypothesis that some milk proteins are expressed in milk not for their intact structure but for the ‘‘encrypted’’ functional peptides that are released during digestion (6). However, whether the previously annotated functional milk peptides are released and present in various segments of the digestive system, the bloodstream, or tissues is mostly unknown. Gastric proteolysis has been presumed to be minimal, with most milk proteins digested in the small bowel (7). This study takes a first step by determining which milk peptides are released in the stomach of the term infant and are thus available in the proximal small bowel.
The full spectrum of peptides released from human milk both before and during digestion by the infant has not been determined. Limitations have been technical: analytical platforms were previously unable to measure low-abundance peptides in complex mixtures. Limitations have also been clinical: obtaining gastric aspirates from breast-fed infants requires placement of a nasogastric tube and often yields very small volumes of sample. This research program was initiated to determine the degree to which biologically relevant milk peptides are available in the proximal gut in vivo.
Milk contains multiple biomolecule classes across a 5-log concentration range within a variety of structures. Breakthroughs in analytical chemistry are making it possible to analyze these milk components in unprecedented detail. Advances in separation science have led to technologies on the nano scale to isolate biomolecules in highly reproducible flow fields (8). Advances in quadrupole time-of-flight (Q-TOF) MS have extended the sensitivity to the subfemtomole level and mass accuracy to low parts per million (9). Advances in computational algorithms allow automated searching through thousands of fragmentation spectra to identify exact peptide sequences and protein of origin (10). Advances in the construction of mass spectral databases are now making it possible to screen entire biomolecule classes on the basis of exact masses and retention times, transforming high-throughput chemistry (11,12). All of these advances were incorporated into a single discovery pipeline platform that is capable of identifying hundreds of naturally occurring unique peptide sequences from submicrogram amounts of sample from actual human infant gastric samples. It is now possible to characterize the proteins and peptides as they are released for their exact sequences, posttranslational modifications, and protein of origin.
The difficulty in obtaining gastric aspirates from healthy infants has led previous studies aimed at characterizing protein digestion in vivo to use animal model surrogates. One study characterized the peptides released in the stomach, jejunum, and ileum of piglets fed an artificial milk formula (13). That study used MS techniques and revealed dozens of peptides in the jejunum and ileum samples.
The aim of the present study was to characterize and compare the entire complement of peptides (peptidome) of intact milk and gastric digestion samples from term infants and to determine whether these peptide sequences are homologous to known functional peptides. This study represents the first time, to our knowledge, in which the in vivo degradation products of milk proteins have been examined extensively. The peptides were then analyzed for existing ontology descriptions for function by using a functional database assembled from the literature.
Participants and Methods
Materials. Acetonitrile, formic acid (FA), and trifluoroacetic acid were obtained from Thermo Fisher Scientific and trichloroacetic acid from EMD Millipore. All water used was nanopure (18.2 ohms).
Participants and samples. This study was approved by the institutional review board of the University of California, Davis (UC Davis). After informed consent, intact milk samples were obtained from 3 healthy mothers who delivered at term (see Table 1 for infant metadata). The breast was cleansed with water on a washcloth (no soap or alcohol), and then the milk samples were collected by an electric pump into a sterile plastic container. Samples from both breasts were combined and then frozen in a home freezer and transported on ice to the Neonatal Intensive Care Unit at the UC Davis Children´s Hospital and kept frozen at -20°C. The time of day of pumping was not specified or recorded. The 3 term infants were hospitalized because of health problems unrelated to the gastrointestinal tract (Table 1). Two of the infants received i.v. antibiotics before sample collection, which may alter the gastric microbiota. None of the infants received oral antibiotics or acid-suppressing agents. The infants' conditions precluded normal feeding; therefore, a nasogastric tube was placed for each. The milk samples were thawed in warm (not hot) water and stored in a refrigerator at 4°C until feeding. Just before feeding, an aliquot of milk was refrozen at -40°C. The unfortified milk was fed via the nasogastric tube over 30 min. Two hours after the initiation of the feeding, a fraction of the gastric contents
was collected back through the tube via suction for each infant and stored at –40°C. Milk and gastric acid samples were transported to the UC Davis laboratory on dry ice and stored at –80°C until analysis.
Sample preparation. Samples were removed from the freezer and thawed on ice. They were then mixed on a vortex for 1 min each. Peptides were extracted as previously described (14), with the following modifications. A 100µL sample of the mother´s expressed breast milk and 100µL of the infant´s gastric fluid were used. To each sample, 100µL of nanopure water were added, and the samples were again mixed on a vortex. The samples were then incubated at 100°C to prevent any further enzyme-driven proteolysis. Briefly, samples were delipidated by centrifugation, and proteins in the infranate samples were precipitated with 1:1 (sample to solution) 200 g/L trichloroacetic acid, followed by mixing on a vortex plate, centrifugation, and removal of the supernatant. Supernatants were applied to a 96-well C18 solid-phase extraction plate to purify peptides and eluted as described (14). Eluted peptides were dried by vacuum centrifugation and rehydrated in 60µL nanopure water for Q-TOF analysis. Two microliters of 0.1 µg/µL peptide standards containing equal parts Leu-enkephalin, gonadoliberin, angiotensin I, and neurotensin (ProteoChem) were added to map the retention time reproducibility of the samples.
Peptide analysis. The analysis was performed with the Agilent nanoliquid chromatography Chip Cube 6520 Q-TOF MS/MS. The chip used contained an enrichment and analytical column packed with a C18 stationary phase. Q-TOF parameters were as described (14). Briefly, the gradient elution solvents were as follows: 3% acetonitrile /0.1% FA (A)
and 90% acetonitrile/0.1% FA (B). The gradient used was ramped from 0% to 8% B from 0 to 5 min, 8% to 26.5% B from 5 to 24 min, 26.5% to 100% B from 24 to 48 min, followed by 100% B for 2 min and 100% A for 10 min (to re-equilibrate the column). Each sample was analyzed in triplicate by using the Q-TOF in MS-only mode.
Creation of a peptide library. To create a peptide library, these gastric and intact samples for term-delivered infants were analyzed by using the same method but with MS/MS as described (14). Data were exported to Mascot Generic Format (.mgf) (14) and analyzed via the downloadable version of X!Tandem (The Global Proteome Machine Organization; thegpm.org) (10) against a library of human milk proteins derived from previous human milk proteomes (15–17). The parameters used in X! Tandem were as previously described (18). Briefly, no complete modifications were required, but possible phosphorylations of serine, threonine, or tyrosine; deamidation of glutamine or asparagine; and oxidation of methionine or tryptophan were allowed. Error tolerances used were 20 ppm for precursor masses and 40 ppm for fragment masses.
The results from X!Tandem for all samples were compiled for milk samples and for gastric samples separately. Peptides with e-values ≤0.01 were removed (a 99% confidence level threshold). To isolate only unique peptide sequences, all duplicates of sequence, protein, and modifications combined were eliminated with the ‘‘remove duplicates’’ function in Excel (Microsoft Corporation). Peptides representing identical amino acid sequences and modifications but modified in different positions were also removed as duplicates. The compiled peptide libraries were then used to make an exclusion list for recursive analysis as described previously (18). Recursive analysis was repeated until the number of new peptides returned for each sample was ≤10. Three rounds of recursive analysis were completed for the milk samples (for a total of 4 MS/MS analyses for each). Five rounds were completed for 2 of the gastric samples (for a total of 6 MS/MS analyses for each). One gastric sample with retention times repeatedly different than the other 2 gastric samples was examined alone, and required 10 rounds of recursive analysis before the number of new peptides returned was #10. Peptides were identified via the above procedure, and the results from the recursive analysis were added to the original peptide results. The data have been deposited to the ProteomeXchange (19) (identifier PXD000688).
Peptide identification. Compounds were identified from all samples by the ‘‘Batch Targeted Feature Extraction’’ function in Agilent MassHunter Profinder B.06.00. The data were searched against the peptide library on the basis of molecular formula. One library generated from milk peptide library was used to extract milk sample peptide peak areas, and 2 different libraries were used to extract the gastric peptides (because 1 sample had consistently different retention times). The milk peptide library could not be applied to the gastric samples because they showed retention time shifts in comparison with the milk peptides, likely due to molecular interactions within the sample matrices. For library searching, the following parameters were used: the maximum number of matches per formula was 1; peaks were matched on both mass (within a 20-ppm error) and retention time (within a 1-min error); a height threshold of 500 ion counts was used; the charge carrier was ‘‘protons,’’ and the allowed charge states were 1–7; and the isotope model was ‘‘peptides.’’ Extracted ion chromatogram peaks were smoothed with the Gaussian function, and the resulting peaks were integrated via the ‘‘agile’’ algorithm. After compounds were extracted, each peak was manually inspected for peak integration, and any incorrect assignments were corrected within the Profinder program.
Statistical analysis. Peptide abundances (peak area) were compared between the milk and gastric samples (n = 3) via paired, 2-tailed t tests. Peptides with a P value ≤0.05 were deemed significantly different.
Functional annotation. Identified peptide sequences from the milk and gastric samples were searched against a library of known functional milk peptides from the literature. Peptides in the samples that completely or partially encompassed the known functional peptide were counted for the bioactive peptide table. For partial matches, peptides had to match the functional peptide in the database by ≥80% overlap. However, in cases in which residues had similar properties (i.e., alanine/valine, isoleucine/valine, aspartic acid/glutamic acid), substitutions were allowed.
Results
The combined base peak chromatograms of the milk peptides in the intact milk and gastric samples are shown in Supplemental Figure 1. The intact milk peptide profiles (in green) contained fewer peaks and peaks that were less abundant than the gastric digestion profiles (in blue). These differences between the 2 groups' base peak chromatograms were the result of the biologic differences, not instrumentation scaling.
The base peak chromatograms for 3 instrumental replicates of a single intact milk sample are shown in Supplemental Figure 2. The extent to which the peaks were overlapping in these replicates demonstrated the high reproducibility of retention time and peak volume of the nano-liquid chromatography–QTOF method. The nano-liquid chromatography chip system used has exhibited highly reproducible retention times (8,20). The low within-sample variation confirms that differences between samples as large as those shown in the base peak chromatogram of Supplemental Figure 1 were due to biologic differences rather than instrumental variation.
Supplemental Figure 3 shows the single mass spectrum for 2 identified compounds (1A: 1189.6343 m/z; 1C: 834.4983 m/z). Supplemental Figure 1B, D shows the tandem spectra for the fragmentation of 1189.6343 m/z and 834.4983 m/z, respectively. Each identified fragment was labeled with the amino acid sequence of the fragment. Both b- and y-type ions are shown in each figure (in black and gray, respectively). These tandem spectra identify the precursor molecules for their amino acid sequences.
Overall, 784 unique naturally occurring peptide sequences were identified on the basis of the database of tandem-identified peptides at 99% confidence. Select peaks were labeled for their sequences and protein of origin. All identified sequences are shown in a table ‘‘Peptide_Table,’’ deposited with the MS/MS spectral files and X!Tandem result files in the ProteomeXchange (http://www.proteomexchange.org; identifier PXD000688). These peptides derived from 36 milk proteins. Each protein had at least 1 unique peptide sequence with an e-value ≤0.01. In the intact milk samples, 198 unique peptides were identified, with an average of 189.8/sample. In the gastric samples, 649 unique peptides were found (average of 418.3/sample), an ∼120% increase over the number found in the intact milk samples (P = 1.2 x 10-6, paired t test). Sixty-four peptides were common to both milk and gastric samples. One hundred thirty-five peptides were present only in the intact milk and were undetectable in the gastric samples. Five hundred eighty-six peptides were present only in the gastric samples. On the basis of abundance, 603 peptides were significantly different between the gastric and the milk samples: 440 were significantly higher in the gastric samples and 163 were significantly lower in the gastric samples (deposited in the ProteomeXchange as ‘‘Statistical_Comparison_Peptides’’).
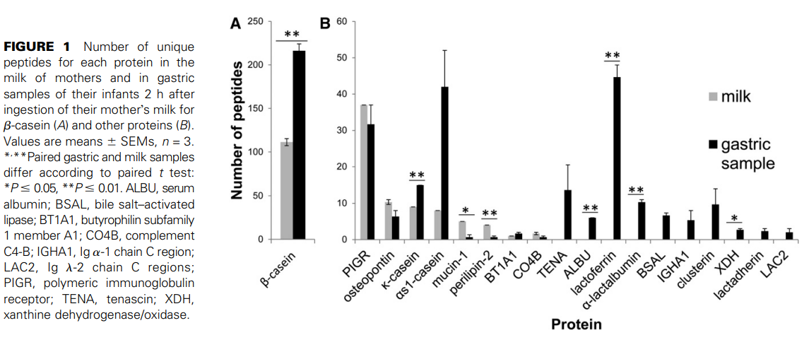
Most peptides identified in the intact milk samples by count were from β-casein (111.5 peptides on average, 59% of the average peptide count). In the gastric samples, on average, 52% of the unique peptide sequences derived from β-casein. This observation suggests that the degradation within milk and the stomach is protein-selective (because b-casein does not represent even half of milk proteins).
In this small sample set, the numbers of unique peptides from some milk proteins such as polymeric immunoglobulin receptor, osteopontin, butyrophilin, tenascin, clusterin, lactadherin, αs1-casein, Ig α-1 chain C region, and Ig λ-2 chain C regions were not significantly different between the intact milk samples and the gastric samples. The number of peptides identified from β-casein, Κ-casein, mucin-1, perilipin-2, serum albumin, lactoferrin, α-lactalbumin, bile salt–activated lipase, and xanthine dehydrogenase/oxidase were significantly different between milk and gastric samples. Peptides released from lactoferrin, α-lactalbumin, serum albumin, xanthine dehydrogenase/oxidase, lactadherin, tenascin, bile salt–activated lipase, Ig α-1 chain C region, and Ig λ-2 chain C regions were not present in the milk but were present as peptides in the gastric samples (Fig. 1). Figure 2 presents the total peptide density by volume (as opposed to number in Fig. 1) from milk proteins, demonstrating that most peptide volume comes from β-casein. The total volumes of peptides from β-casein, lactoferrin, and osteopontin were significantly higher in gastric samples than in milk samples. The number of precursor proteins identified (proteins that yielded measurable peptides) was lower in milk compared with gastric samples (11 vs. 35 proteins identified in intact milk and gastric digesta, respectively, P < 0.05).
Of the 784 peptides identified here, 42 were phosphorylated at serine, threonine, or tyrosine. Phosphorylated peptides made up 5.4% of the total peptides. Twenty-eight phosphorylated peptides were present in the milk and 19 in the gastric samples. Most of these phosphopeptides are derived from the casein proteins, which are known to be multiply phosphorylated. Indeed, several phosphopeptides identified were multiply phosphorylated: 16 were doubly phosphorylated and 7 were triply phosphorylated. Milk peptides with multiple phosphorylation are known to bind calcium and may aid in calcium delivery (3).
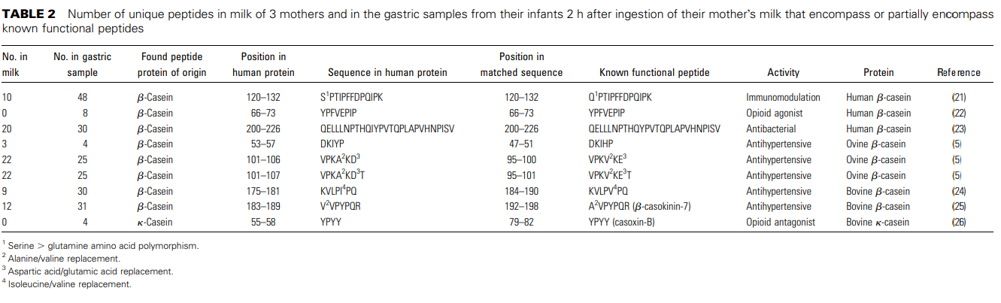
The peptides were compared against a database of functional peptides from the literature. Ninety-eight peptides in milk and 205 peptides in the gastric samples were identified as entirely or partially encompassing known functional peptides (Table 2). The peptides found in the milk samples matched published antibacterial, immunomodulatory, and antihypertensive peptide sequences. Peptides in the gastric sample matched sequences with the same functions plus opioid antagonist and opioid agonist activities. All of the functional database peptides that mapped to the peptides identified here were derived from human b-casein, bovine b-casein, ovine b-casein, and bovine k-casein. It is not clear whether the lack of functional annotation for the remaining peptides is due to lack of biologic activity or lack of scientific study.
Discussion
This study used a novel, comprehensive, and highly sensitive analytical pipeline to identify hundreds of naturally occurring peptides from human milk and peptides released in the term infant stomach. Hundreds of milk protein fragments created by in vitro degradation have demonstrated biologic activity; however, at what anatomic location these functional peptides appear during infant digestion has been unclear. This study represents a first step by mapping milk protein degradation in the term infant stomach and suggests that many functional peptides are biologically available in the proximal gut.
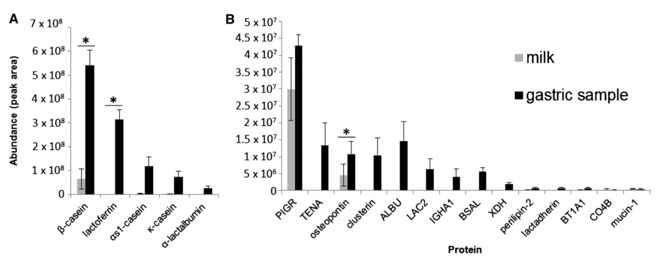
FIGURE 2 Total volume for all peptides by protein in the milk of mothers and in gastric samples of their infants 2 h after ingestion of their mother´s milk for β-casein, lactoferrin, αs1-casein, k-casein, and α-lactalbumin (A) and other proteins (B). Values are means ± SEMs, n = 3.
*Paired gastric and milk samples differ according to paired t test, P ≤ 0.05. ALBU, serum albumin; BSAL, bile salt–activated lipase; BT1A1, butyrophilin subfamily 1 member A1; CO4B, complement C4-B; IGHA1, Ig a-1 chain C region; LAC2, Ig l-2 chain C regions; PIGR, polymeric immunoglobulin receptor; TENA, tenascin; XDH, xanthine dehydrogenase/oxidase.
The large number of peptides in the milk samples suggests that proteolysis begins in the breast itself during lactation or milk let-down and/or that significant proteolysis occurred between the time of milk expression and feeding (i.e., during collection, freezing, thawing, or refrigeration of the samples). The 64 peptides found in both intact and gastric samples may have structural resistance to gastric degradation. The 135 peptides found only in the intact milk were likely degraded in the stomach.
The increase in unique peptides from highly abundant milk proteins in the stomach (lactoferrin, a-lactalbumin, bile salt–activated lipase, k-casein, and serum albumin) that were absent in the milk suggests that extensive proteolysis occurs in the term infant stomach. This finding has clinical relevance because the antibacterial, immunomodulatory, and calcium-binding functions of these peptides are particularly relevant in the small bowel. The small bowel has predominately digestive andabsorptive functions and is not as well suited to protect the epithelial monolayer from luminal bacteria as the colon with its thick mucus layer and lack of villi. It is possible that release of human milk peptides in the very proximal gut plays an important role in the decreased incidence of diarrheal disease in breast-fed infants. It is likely that the premature infant differs from the term infant in gastric proteolysis of human milk; if this is the case, these differences may contribute to the observed differences in the fecal microbiota, in growth, and in susceptibility to sepsis, necrotizing enterocolitis, and metabolic bone disease between term and preterm infants.
This study has several limitations. First, the small sample size and differences in day after birth of collection of samples limit the statistical analysis. For instance, it appears that there is a trend toward greater numbers of peptides in the samples from the infant obtained at day 12 compared with the infant whose samples were obtained at day 4. Serial samples over time from the same mother-infant dyad would be helpful to address this hypothesis. Second, because these infants were tube-fed, effects of infant oral enzymes were bypassed, except for those due to swallowed saliva. Because humans are known to secrete several antiproteases, but no proteases, in saliva (6), it is possible that direct breastfeeding or bottle feeding of expressed breast milk may be associated with less protein degradation than observed here. Likewise, bypassing the oral cavity may affect gastric enzyme secretion. In adult humans, the cephalic phase of digestion (sight, smell, taste, thought) causes an increase in gastric acid secretion along with an increase in chymotrypsin, trypsin, and lipase concentrations (27); however, pepsin release is not consistently observed (28). Whether human infants have similar responses is unknown. Third, because the milk samples were not sterilized, it is possible that bacteria in the milk and bacteria in the infant stomach could alter digestion of milk proteins either directly or indirectly. Because most protein cleavages are due to the milk enzyme plasmin, it seems likely that direct bacterial protein digestion is minimal. Furthermore, in a previous study comparing peptides from boiled and unheated milk samples, we found 96% identical results (18), suggesting that bacterial digestion of milk proteins is minimal.
The known milk proteins originate from genes that are widely dispersed across the genome (29). Annotation of functions must now account for a broader array of biologic high-throughput approach used for this study identified hundreds of unique peptide sequences from mothers´ milk samples and milk after digestion for 2 h in the infant stomach. The chipbased MS approach with database peptide identification successfully characterized the digestion of milk in the infant´s stomach. This work is the first, to our knowledge, to characterize human digestion at a peptidomic level. The validity of this methodologic platform for peptidomic profiling is now established and can now be used for larger studies exploring protein degradation across digestion.
Acknowledgments
The authors thank Cora J. Dillard for editing this manuscript and Jose Meza from Agilent Technologies for technical support. D.C.D., J.B.G., D.B., and M.A.U. designed the research; R.B. collected clinical samples; D.C.D., A.G., and C.B.L. established the peptidomics methodology; D.C.D. performed the MS analysis; D.C.D., A.B., N.K., and A.G. analyzed the data; and D.C.D. wrote the manuscript and had primary responsibility for the final content. All authors read and approved the final manuscript.
Literature Cited
- Liepke C, Zucht HD, Forssmann WG, Standker L. Purification of novel peptide antibiotics from human milk. J Chromatogr B Biomed Sci Appl.2001;752:369–77.
- Politis I, Chronopoulou R. Milk peptides and immune response in the neonate. In: Bo¨sze Z, editor. Bioactive components of milk. Berlin: Springer-Verlag; 2008. p. 253–69.
- Sato R, Noguchi T, Naito H. Casein phosphopeptide (CPP) enhances calcium absorption from the ligated segment of rat small intestine. J Nutr Sci Vitaminol (Tokyo). 1986;32:67–76.
- Liepke C, Adermann K, Raida M, Ma¨gert HJ, Forssmann WG, Zucht HD. Human milk provides peptides highly stimulating the growth of bifidobacteria. Eur J Biochem. 2002;269:712–8.
- Gomez-Ruiz J ´ A, Ramos M, Recio I. Angiotensin-converting enzyme- ´inhibitory peptides in Manchego cheeses manufactured with different starter cultures. Int Dairy J. 2002;12:697–706.
- Dallas DC, Underwood MA, Zivkovic AM, German JB. Digestion of protein in premature and term infants. J Nutr Disord Ther. 2012;2:1–9.
- Britton JR, Koldovsky O. Development of luminal protein digestion: implications for biologically active dietary polypeptides. J Pediatr Gastroenterol Nutr. 1989;9:144.
- Chu CS, Nin˜onuevo MR, Clowers BH, Perkins PD, An HJ, Yin H, Killeen K, Miyamoto S, Grimm R, Lebrilla CB. Profile of native N-linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics. 2009;9:1939–51.
- Nin˜onuevo M, An H, Yin H, Killeen K, Grimm R, Ward R, German B, Lebrilla C. Nanoliquid chromatography mass spectrometry of oligosaccharides employing graphitized carbon chromatography on microchip with a high accuracy mass analyzer. Electrophoresis. 2005;26:3641–9.
- Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–7.
- Aldredge D, An HJ, Tang N, Waddell K, Lebrilla CB. Annotation of a serum N-glycan library for rapid identification of structures. J Proteome Res. 2012;11:1958–68.
- Wu S, Grimm R, German JB, Lebrilla CB. Annotation and structural analysis of sialylated human milk oligosaccharides. J Proteome Res.2011;10:856–68.
- Bouzerzour K, Morgan F, Cuinet I, Bonhomme C, Jardin J, Le Hue¨rouLuron I, Dupont D. In vivo digestion of infant formula in piglets: protein digestion kinetics and release of bioactive peptides. Br J Nutr. 2012;108:2105–14.
- Dallas DC, Guerrero A, Parker EA, Garay LA, Bhandari A, Lebrilla CB, Barile D, German JB. Peptidomic profile of milk of Holstein cows at peak lactation. J Agric Food Chem. 2013;62(1):58–65.
- Molinari CE, Casadio YS, Hartmann BT, Livk A, Bringans S, Arthur PG, Hartmann PE. Proteome mapping of human skim milk proteins in term and preterm milk. J Proteome Res. 2012;11(3):1696–714.
- Mange A, Bellet V, Tuaillon E, Van de Perre P, Solassol J. Comprehen- ´sive proteomic analysis of the human milk proteome: contribution of protein fractionation. J Chromatogr B Analyt Technol Biomed Life Sci.2008;876:252–6.
- Liao Y, Alvarado R, Phinney B, Lo¨nnerdal B. Proteomic characterization of human milk fat globule membrane proteins during a 12 month lactation period. J Proteome Res. 2011;10:3530–41.
- Dallas DC, Guerrero A, Khaldi N, Castillo PA, Martin WF, Smilowitz JT, Bevins CL, Barile D, German JB, Lebrilla CB. Extensive in vivo human milk peptidomics reveals specific proteolysis yielding protective antimicrobial peptides. J Proteome Res. 2013;12:2295–304.
- Vizca´ıno JA, Coˆte RG, Csordas A, Dianes JA, Fabregat A, Foster JM, ´Griss J, Alpi E, Birim M, Contell J. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–9.
- Tao N, DePeters EJ, German JB, Grimm R, Lebrilla CB. Variations in bovine milk oligosaccharides during early and middle lactation stages analyzed by high-performance liquid chromatography-chip/mass spectrometry. J Dairy Sci. 2009;92:2991–3001.
- Azuma N, Nagaune S, Ishino Y, Mori H, Kaminogawa S, Yamauchi K.DNA-synthesis stimulating peptides from human b-casein. Agric Biol Chem. 1989;53:2631–4.
- Meisel H, FitzGerald R. Opioid peptides encrypted in intact milk protein sequences. Br J Nutr. 2000;84:S27–31.
- Hayes M, Stanton C, Fitzgerald GF, Ross RP. Putting microbes to work: dairy fermentation, cell factories and bioactive peptides. Part II: bioactive peptide functions. Biotechnol J. 2007;2:435–49.
- Maruyama S, Nakagomi K, Tomizuka N, Suzuki H. Angiotensin I-converting enzyme inhibitor derived from an enzymatic hydrolysate of casein. II. Isolation and bradykinin-potentiating activity on the uterus and the ileum of rats. Agric Biol Chem. 1985;49:1405–9.
- Clare DA, Swaisgood HE. Bioactive milk peptides: a prospectus. J Dairy Sci. 2000;83:1187–95.
- Chiba H, Tani F, Yoshikawa M. Opioid antagonist peptides derived from kappa-casein. J Dairy Res. 1989;56:363–6.
- Mattes RD. Physiologic responses to sensory stimulation by food: nutritional implications. J Am Diet Assoc. 1997;97:406–13.
- Brand JG, Cagan R, Naim M. Chemical senses in the release of gastric and pancreatic secretions. Annu Rev Nutr. 1982;2:249–76.
- Lemay DG, Lynn D, Martin W, Neville M, Casey T, Rincon G, Kriventseva E, Barris W, Hinrichs A, Molenaar A. The bovine lactation genome: insights into the evolution of mammalian milk. Genome Biol. 2009;10:R43.